You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002378_00193
You are here: Home > Sequence: MGYG000002378_00193
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Akkermansia sp001580195 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Verrucomicrobiales; Akkermansiaceae; Akkermansia; Akkermansia sp001580195 | |||||||||||
| CAZyme ID | MGYG000002378_00193 | |||||||||||
| CAZy Family | GH109 | |||||||||||
| CAZyme Description | Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 11432; End: 12709 Strand: - | |||||||||||
Full Sequence Download help
| MGAPIWNRRN FLKTAAGTAG FLMAAPYMNA AESLTDARPL KVGLVGCGGR GLGAMKNALD | 60 |
| ADPGTVVWAL ADVFEERTGF GARLLTEQYG PRMQADKSRM FHGLDGYRKL LQTDIDVVLL | 120 |
| CTPPAFRPEH LAAAIDANKH IYAEKPVAVD VPGVLSVLES ARLAKLKNLV ILDGFCWRYD | 180 |
| KANEEAHRKL ASGELGRVLS FDGLYYTTPP KSPLALDSRP ASDTDVAWAL RNWTAWNWLS | 240 |
| GGQFVEQIVH TIDGMVWSMN EQLPLAAFGS GGRAQRRDDG DVWDHYDVYF EYDHNVVGHI | 300 |
| SCRQWVGCHG EITDRTVCEK GMLVTPYRPH IQADKRWRFR GERGNMYADT HVAFYRFLRS | 360 |
| GTWTQTLESA ANKTLVAILG REAAHTGQRI TWEQIKKNTA RLVPEDLTMD TPLPPARIPR | 420 |
| PGRTA | 425 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH109 | 40 | 205 | 1.9e-16 | 0.39598997493734334 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0673 | MviM | 1.54e-33 | 38 | 386 | 2 | 321 | Predicted dehydrogenase [General function prediction only]. |
| pfam01408 | GFO_IDH_MocA | 3.76e-10 | 40 | 171 | 1 | 117 | Oxidoreductase family, NAD-binding Rossmann fold. This family of enzymes utilize NADP or NAD. This family is called the GFO/IDH/MOCA family in swiss-prot. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QWO92521.1 | 0.0 | 1 | 425 | 1 | 425 |
| QWP69861.1 | 0.0 | 1 | 425 | 1 | 425 |
| QWO85217.1 | 0.0 | 1 | 425 | 1 | 425 |
| QWO95053.1 | 0.0 | 1 | 425 | 1 | 425 |
| QWO89672.1 | 0.0 | 1 | 425 | 1 | 425 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3RBV_A | 3.47e-13 | 32 | 211 | 20 | 182 | CrystalStructure of KijD10, a 3-ketoreductase from Actinomadura kijaniata incomplex with NADP [Actinomadura kijaniata],3RC1_A Crystal Structure of KijD10, a 3-ketoreductase from Actinomadura kijaniata incomplex with NADP and TDP-benzene [Actinomadura kijaniata],3RC2_A Crystal Structure of KijD10, a 3-ketoreductase from Actinomadura kijaniata in complex with TDP-benzene and NADP; open conformation [Actinomadura kijaniata] |
| 3RC7_A | 3.47e-13 | 32 | 211 | 20 | 182 | CrystalStructure of the Y186F mutant of KijD10, a 3-ketoreductase from Actinomadura kijaniata in complex with TDP-benzene and NADP [Actinomadura kijaniata] |
| 3RCB_A | 1.12e-12 | 32 | 211 | 20 | 182 | Crystalstructure of the K102E mutant of KijD10, a 3-ketoreductase from Actinomadura kijaniata in complex with TDP-benzene and NADP [Actinomadura kijaniata] |
| 3RC9_A | 2.00e-12 | 32 | 211 | 20 | 182 | CrystalStructure of the K102A mutant of KijD10, a 3-ketoreductase from Actinomadura kijaniata in complex with TDP-benzene and NADP [Actinomadura kijaniata] |
| 3EC7_A | 1.21e-10 | 40 | 232 | 24 | 207 | CrystalStructure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_B Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_C Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_D Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_E Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_F Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_G Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_H Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B3TMR8 | 1.67e-12 | 32 | 211 | 2 | 164 | dTDP-3,4-didehydro-2,6-dideoxy-alpha-D-glucose 3-reductase OS=Actinomadura kijaniata OX=46161 GN=KijD10 PE=1 SV=1 |
| A9N564 | 1.85e-10 | 40 | 261 | 3 | 187 | Inositol 2-dehydrogenase OS=Salmonella paratyphi B (strain ATCC BAA-1250 / SPB7) OX=1016998 GN=iolG PE=3 SV=1 |
| B5F3F4 | 1.85e-10 | 40 | 261 | 3 | 187 | Inositol 2-dehydrogenase OS=Salmonella agona (strain SL483) OX=454166 GN=iolG PE=3 SV=1 |
| Q8ZK57 | 1.85e-10 | 40 | 261 | 3 | 187 | Inositol 2-dehydrogenase OS=Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) OX=99287 GN=iolG PE=1 SV=1 |
| Q9ALN5 | 5.86e-09 | 38 | 209 | 3 | 158 | dTDP-3,4-didehydro-2,6-dideoxy-alpha-D-glucose 3-reductase OS=Saccharopolyspora spinosa OX=60894 GN=spnN PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as TAT
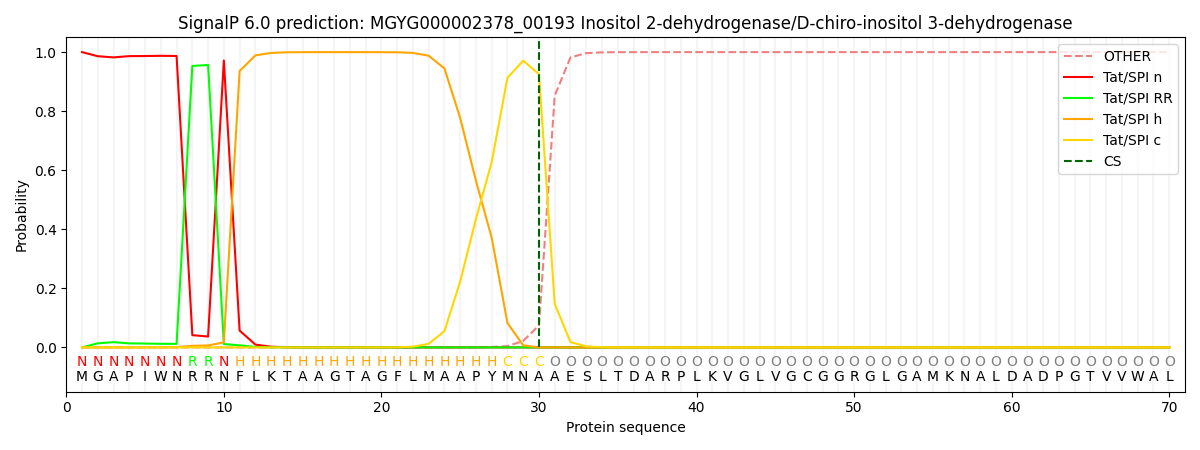
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 0.000000 | 0.999971 | 0.000000 | 0.000000 |
