You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002564_00001
You are here: Home > Sequence: MGYG000002564_00001
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-831 sp902388455 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA932; CAG-831; CAG-831 sp902388455 | |||||||||||
| CAZyme ID | MGYG000002564_00001 | |||||||||||
| CAZy Family | GH110 | |||||||||||
| CAZyme Description | Alpha-1,3-galactosidase B | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 225; End: 1985 Strand: + | |||||||||||
Full Sequence Download help
| MKRIVITLLL LSIIRPALEG RIFEMSAFGI RPGATNLPEK ISSALESIRK ETGHGDSVTL | 60 |
| RFLPGRYDFH STDALQRTYY ISNHDQSEQQ AVGIAIEGWE NLTLDGRGAE LVFHGVIIPF | 120 |
| AVIDSKGCSV KNLSIDFENP QTAQVRIIEN RGAGGIVFRP ESWVKYRISQ DSIFEVYGEG | 180 |
| WALRPSTGIA FEPDTRHILW NTGDLFYSTA GVRKTEHGLL APQWQDSRLT PGTVVAMRTW | 240 |
| SRPAPGIFLY GNTDTRLTNI NVHYAFGMGL IAQFCDGITL NGFSVCLKNE NDPRYFTTQA | 300 |
| DATHFSQCSG IIQSSGGLYE GMMDDAINVH GVYLKVTGVR GDRSITARFM HDQAWGFRWG | 360 |
| CPGDSVRMLN ADVMEYTGSL LHIESITPDN TGHGSKELII TFREPVEIDP ERMNIGIENL | 420 |
| SRTPEVIFSG NTVRNNRARG ALFSSPRRTV VENNVFDHTS GSAILLCGDC NGWYESGACR | 480 |
| DILIKDNRFI NALTSMYQFT TAVISIYPEI PDLHNQTVYF HGGHPGAITI TGNTFETFDM | 540 |
| PVLYAKSVDG LVFKDNTVIV NNDYRPFHKN RNRFLLERTD NIEISY | 586 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH110 | 24 | 585 | 1.8e-192 | 0.9908759124087592 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRO24974.1 | 2.74e-259 | 9 | 585 | 10 | 591 |
| EDY94243.1 | 7.12e-250 | 16 | 585 | 17 | 591 |
| ABR37834.1 | 2.35e-248 | 21 | 585 | 22 | 591 |
| QUT56908.1 | 2.35e-248 | 21 | 585 | 22 | 591 |
| QQY40418.1 | 2.52e-248 | 21 | 585 | 24 | 593 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7JW4_A | 2.92e-71 | 17 | 585 | 22 | 593 | Crystalstructure of PdGH110B in complex with D-galactose [Pseudoalteromonas distincta],7JW4_B Crystal structure of PdGH110B in complex with D-galactose [Pseudoalteromonas distincta] |
| 7JWF_A | 1.53e-70 | 17 | 585 | 22 | 593 | Crystalstructure of PdGH110B D344N in complex with alpha-(1,3)-galactobiose [Pseudoalteromonas distincta],7JWF_B Crystal structure of PdGH110B D344N in complex with alpha-(1,3)-galactobiose [Pseudoalteromonas distincta],7JWF_C Crystal structure of PdGH110B D344N in complex with alpha-(1,3)-galactobiose [Pseudoalteromonas distincta],7JWF_D Crystal structure of PdGH110B D344N in complex with alpha-(1,3)-galactobiose [Pseudoalteromonas distincta] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A6KWM0 | 4.69e-249 | 21 | 585 | 22 | 591 | Alpha-1,3-galactosidase B OS=Phocaeicola vulgatus (strain ATCC 8482 / DSM 1447 / JCM 5826 / CCUG 4940 / NBRC 14291 / NCTC 11154) OX=435590 GN=glaB1 PE=3 SV=1 |
| Q5LGZ8 | 1.90e-239 | 8 | 585 | 12 | 594 | Alpha-1,3-galactosidase B OS=Bacteroides fragilis (strain ATCC 25285 / DSM 2151 / CCUG 4856 / JCM 11019 / NCTC 9343 / Onslow) OX=272559 GN=glaB PE=1 SV=1 |
| Q64XV2 | 3.82e-239 | 8 | 585 | 12 | 594 | Alpha-1,3-galactosidase B OS=Bacteroides fragilis (strain YCH46) OX=295405 GN=glaB PE=3 SV=1 |
| A6L2M8 | 1.19e-230 | 1 | 584 | 1 | 590 | Alpha-1,3-galactosidase B OS=Phocaeicola vulgatus (strain ATCC 8482 / DSM 1447 / JCM 5826 / CCUG 4940 / NBRC 14291 / NCTC 11154) OX=435590 GN=glaB2 PE=3 SV=1 |
| A6LFT2 | 8.14e-213 | 23 | 584 | 26 | 588 | Alpha-1,3-galactosidase B OS=Parabacteroides distasonis (strain ATCC 8503 / DSM 20701 / CIP 104284 / JCM 5825 / NCTC 11152) OX=435591 GN=glaB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
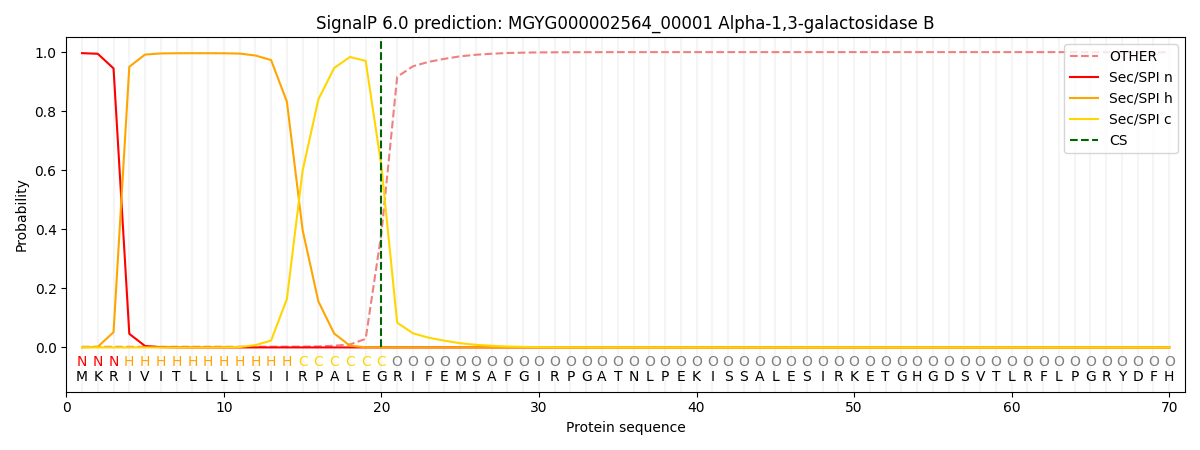
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.005263 | 0.991522 | 0.002392 | 0.000276 | 0.000237 | 0.000261 |
