You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000764_00043
You are here: Home > Sequence: MGYG000000764_00043
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Porphyromonas_A sp900541275 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Porphyromonadaceae; Porphyromonas_A; Porphyromonas_A sp900541275 | |||||||||||
| CAZyme ID | MGYG000000764_00043 | |||||||||||
| CAZy Family | GH123 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 55772; End: 57526 Strand: - | |||||||||||
Full Sequence Download help
| MRKRITALLA GLLSLTTAYG QYPVESYQEL PNPTPTDRSL WESLSAPVIG WGSIDIRYPK | 60 |
| ERPADGLTSK EIRLSGWRGE RVYAQAVVSS ATPLTALSYE ISELRSCDGA VIPSPQGESG | 120 |
| SGFVRYVLTD ELNKDGLGGC GERPDHSLFD STLVADGIDH LARSLDLTPC TTRPIWFSID | 180 |
| IPRDARAGSY RATVTVRNGD RTLATLPLSV VVDERALPEK SDFFLDLWQN PYSIARYYGV | 240 |
| TPWSEEHFSI MRPYMEMYRD AGGKVITASI IYKPWNGQTH DPFDSMIDWR LGRDGKWHFD | 300 |
| YTHFDEWVSF MMGLGIDRAI TCFSMVPWEL SFRYFDEASG EYKYLKAEPG SEPYTELWSA | 360 |
| MLRSFAQHLR TKGWNDRTYI AMDERSPEVM EECFRLIQEA DPELKVHMAG ALHEELSDHL | 420 |
| DYYCVGLAAK YSEELKAKRR SEGKITTFYT CCAEPRPNTF TFSAAADGEW YGWYAAREGL | 480 |
| DGYLRWAYNS WVEQPLLDSR YSNWAAGDTY LVYPGVRSSI RFERLRRGIQ AFDKIRALRH | 540 |
| QYAGDRQALA KIDEALSLFD ESKLSPSYTS DRIINEAREI IETL | 584 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH123 | 49 | 582 | 2.5e-202 | 0.9851301115241635 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13320 | DUF4091 | 8.42e-23 | 479 | 539 | 1 | 66 | Domain of unknown function (DUF4091). This presumed domain is functionally uncharacterized. This domain family is found in bacteria, archaea and eukaryotes, and is approximately 70 amino acids in length. There is a single completely conserved residue G that may be functionally important. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM53703.1 | 4.17e-232 | 1 | 581 | 4 | 603 |
| AVM58348.1 | 1.11e-229 | 1 | 580 | 4 | 602 |
| AEW22295.1 | 8.49e-222 | 6 | 581 | 23 | 603 |
| BAR50673.1 | 1.20e-221 | 6 | 581 | 23 | 603 |
| ADV44431.1 | 2.50e-221 | 2 | 580 | 5 | 603 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5L7V_A | 3.86e-211 | 25 | 581 | 4 | 560 | ChainA, glycoside hydrolase [Phocaeicola vulgatus ATCC 8482],5L7V_B Chain B, glycoside hydrolase [Phocaeicola vulgatus ATCC 8482] |
| 5L7R_A | 4.58e-211 | 20 | 581 | 14 | 575 | ChainA, glycoside hydrolase [Phocaeicola vulgatus ATCC 8482],5L7R_B Chain B, glycoside hydrolase [Phocaeicola vulgatus ATCC 8482],5L7U_A Chain A, Glycoside hydrolase [Phocaeicola vulgatus ATCC 8482],5L7U_B Chain B, Glycoside hydrolase [Phocaeicola vulgatus ATCC 8482] |
| 5FQE_A | 4.38e-71 | 158 | 538 | 130 | 542 | Thedetails of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens [Clostridium perfringens],5FQE_B The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens [Clostridium perfringens],5FQF_A The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens [Clostridium perfringens],5FQF_B The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens [Clostridium perfringens],5FR0_A The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens [Clostridium perfringens] |
| 5FQG_A | 6.24e-70 | 158 | 538 | 130 | 542 | Thedetails of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens [Clostridium perfringens],5FQH_A The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens [Clostridium perfringens] |
SignalP and Lipop Annotations help
This protein is predicted as SP
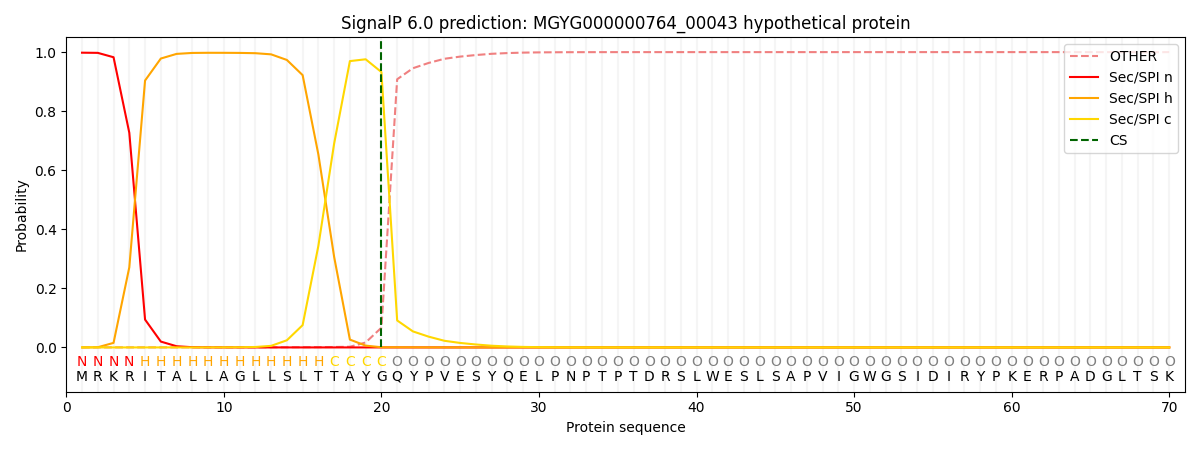
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001560 | 0.995311 | 0.002418 | 0.000277 | 0.000223 | 0.000195 |
