You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000222_00073
You are here: Home > Sequence: MGYG000000222_00073
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parabacteroides sp003473295 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides sp003473295 | |||||||||||
| CAZyme ID | MGYG000000222_00073 | |||||||||||
| CAZy Family | GH127 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 97366; End: 99237 Strand: - | |||||||||||
Full Sequence Download help
| MNAASVVKGL SIIFFCSCIL SGTAQNNSTS SVKHVKISGY IGNRIDDCIE HRVKAQDVEH | 60 |
| LVEPFRHHEE KNLWQSEFWG KWIQGAIASY RYNQDAELYK IISDAAESLM ATQQENGYIG | 120 |
| NYAPEYQLQQ WDIWGRKYTS LGLIAWYDLS GDKKALDAAC RVIDHLMTQV GPQKVNIVTT | 180 |
| GNYVGMPSSS ILEPVMYLYN RTKEQRYLDF ANYIAEQWET PEGPQLISKA IADIPVANRF | 240 |
| PHPESWFSRE NGQKAYEMMS CYEGLLELYK VTGNPLYLTV VEKTVGHIIR EEINIAGSGS | 300 |
| AFECWYGGHD RQTQPTYHTM ETCVTFTWMQ LCNRLLQMTG NSLYADYMEK TIYNALMASL | 360 |
| KADASQIAKY SPLEGWRHEG EEQCGMHINC CNANGPRAFA MIPQFAYQVK EARINVNFYA | 420 |
| PSEAEIVLPG KKEISLKQLT DYPATDKIEL EVSPVKETVC TIALRIPAWS TIAEVTINGQ | 480 |
| AEKGVLQGCY FPVTRKWKKG DKITLQLDLR ARLIEQNQAQ AIVRGPLVLA RDSRFNDGFV | 540 |
| DETSVIVSKD GFVELTPVEV PAFAWLAFTA PMILGTDLEG DRTPRQIHFC DFASAGNTWD | 600 |
| KLQRYRVWLP KTLNVMNSPY KPY | 623 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH127 | 68 | 529 | 1.9e-63 | 0.9103053435114504 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07944 | Glyco_hydro_127 | 1.55e-65 | 52 | 530 | 42 | 503 | Beta-L-arabinofuranosidase, GH127. One member of this family, from Bidobacterium longicum, UniProtKB:E8MGH8, has been characterized as an unusual beta-L-arabinofuranosidase enzyme, EC:3.2.1.185. It rleases l-arabinose from the l-arabinofuranose (Araf)-beta1,2-Araf disaccharide and also transglycosylates 1-alkanols with retention of the anomeric configuration. Terminal beta-l-arabinofuranosyl residues have been found in arabinogalactan proteins from a mumber of different plantt species. beta-l-Arabinofuranosyl linkages with 1-4 arabinofuranosides are also found in the sugar chains of extensin and solanaceous lectins, hydroxyproline (Hyp)2-rich glycoproteins that are widely observed in plant cell wall fractions. The critical residue for catalytic activity is Glu-338, in a ET/SCAS sequence context. |
| COG3533 | COG3533 | 4.02e-65 | 24 | 609 | 6 | 585 | Uncharacterized conserved protein, DUF1680 family [Function unknown]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ARE60508.1 | 1.27e-236 | 28 | 623 | 20 | 620 |
| AGB41651.1 | 3.31e-42 | 81 | 554 | 76 | 584 |
| ADH61394.1 | 4.55e-42 | 32 | 536 | 14 | 565 |
| SNY90695.1 | 2.08e-41 | 31 | 580 | 23 | 601 |
| QOY86860.1 | 5.48e-41 | 81 | 528 | 97 | 547 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6EX6_A | 8.66e-33 | 81 | 536 | 74 | 554 | TheGH127, Beta-arabinofuranosidase, BT3674 [Bacteroides thetaiotaomicron VPI-5482],6EX6_B The GH127, Beta-arabinofuranosidase, BT3674 [Bacteroides thetaiotaomicron VPI-5482] |
| 4QJY_A | 2.27e-28 | 47 | 528 | 40 | 558 | Crystalstructure of native Ara127N, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus],4QJY_B Crystal structure of native Ara127N, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus] |
| 4QK0_A | 3.96e-26 | 47 | 528 | 40 | 558 | Crystalstructure of Ara127N-Se, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus],4QK0_B Crystal structure of Ara127N-Se, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus],4QK0_C Crystal structure of Ara127N-Se, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus],4QK0_D Crystal structure of Ara127N-Se, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus] |
| 5MQO_A | 1.76e-23 | 81 | 533 | 105 | 610 | Glycosidehydrolase BT_1003 [Bacteroides thetaiotaomicron] |
| 6YQH_AAA | 4.32e-21 | 321 | 530 | 337 | 560 | ChainAAA, Acetyl-CoA carboxylase, biotin carboxylase [Bacteroides thetaiotaomicron VPI-5482] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| E8MGH8 | 3.91e-18 | 81 | 538 | 80 | 574 | Non-reducing end beta-L-arabinofuranosidase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=hypBA1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
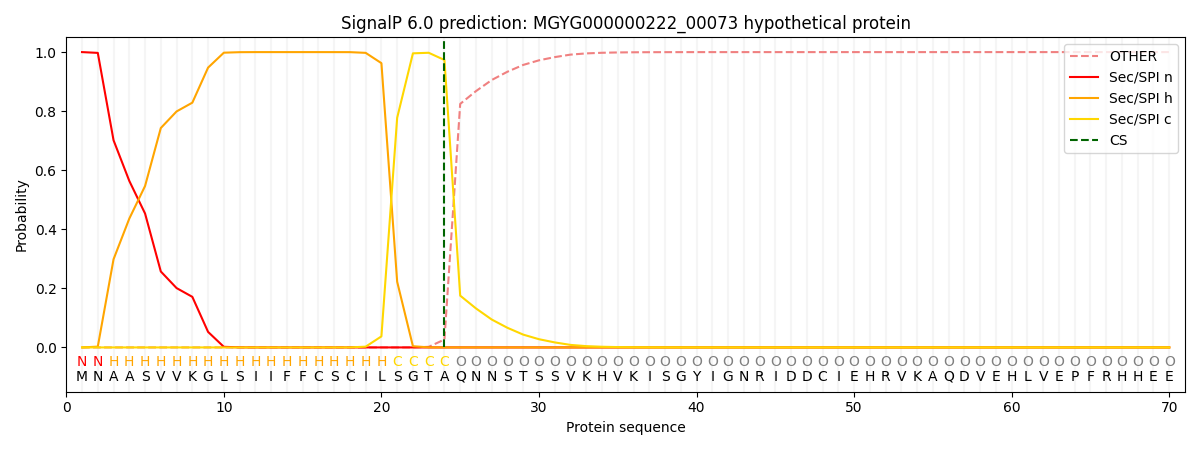
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000244 | 0.999147 | 0.000182 | 0.000143 | 0.000135 | 0.000123 |
