You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001014_00049
You are here: Home > Sequence: MGYG000001014_00049
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
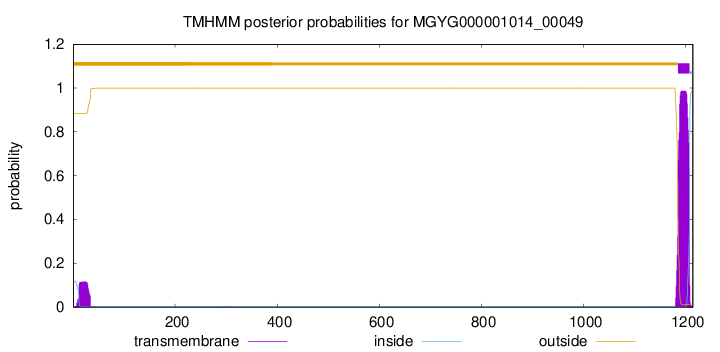
TMHMM annotations
Basic Information help
| Species | Varibaculum timonense | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Actinomycetaceae; Varibaculum; Varibaculum timonense | |||||||||||
| CAZyme ID | MGYG000001014_00049 | |||||||||||
| CAZy Family | GH136 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 890; End: 4534 Strand: + | |||||||||||
Full Sequence Download help
| MNLKKLAKAT GITMLATSLV ALSQLPAVAA PVQAEIIPAT TYYVSAAGND ANDGKSEDKP | 60 |
| LKTLDAVNKL TLHAGDKILL KRGDVFNNQH LCISGDKAFK FNDGTTCAYQ ADSDGAPLVI | 120 |
| SAYGEGDKPV IAANGQGQWK LDYGLTLGHS QHYLAKDAVS STVMLKDAEN IELSDLEITN | 180 |
| KRIEDGSGKE NGCAYNDNCA LDRTGIAGIA QNQGTLDHVV LRNLYVHDVK GNIYNKHSVN | 240 |
| GAIYFSSLIA SDQMKPAADR TAPGFKPALD KPEKGYPRFN DLQIINNRVE NSGRWGIAAV | 300 |
| YSAYASVAVD GRIQIPDEVI TKYGSTNVVI SNNFIKNIGG DAITAMYGYK PVVESNVATE | 360 |
| IATQMNTTDY LFAKDTPGKF AKNGGRVAAS IWPWKSKDAV FRFNEGFNTQ NADKGNGDGM | 420 |
| PWDADSGDGT VYEYNYSANN SGGTVMFCGD QAANSTFRFN IAQNDSLGAL SPTFWDGGYS | 480 |
| GKYPNAHVYN NTFYLKAGSS ILHPAQAQQG TMKVENNIFY NMSDQPRTEA WNPLGHITWD | 540 |
| PKDIDGNAKV AYSHNLYYNY ANYPASDLNP VKVSKGSAVL ADPGKAPSDA ATDMKARPHY | 600 |
| FPGLSYAEAD AKTTVFDGYK LAANSPAIGK GKEIVDDNGF ALGTDFFGRT ANTFDIGAVS | 660 |
| TNQKPLPPAT VTDSAKLVST TYQVTDGDTV PSKNGAYKAG NTIHVPSTEK SPSTIAEVLS | 720 |
| NLTVEGKAQI VDAAGNAQAE DALVAAGMKL KITSLDGKVT SEFPLSIDNV YSWLDDYTHN | 780 |
| VQGNVWFGQK QTGANGKWVN IDKADPAWPN WQVDEYYGPG INGAQLAPTD RSALHGLLSD | 840 |
| SPTSAGATAM AWRAPKTGYI TFKVKDAAAG KPAEPFLRQG DATGGTVTVK LLVNGIEKRS | 900 |
| LTLAKKDIRP DGEFNFTAND ANALQVSKGD FVRVVAESTG TPAQPSMFVS PTITYIDRTV | 960 |
| DTEVIPVKPG TDTGEPDTPT PGQNPQTDKG YVEGKVPGFA LSLAGDKVTG KVTLPQVEGI | 1020 |
| EYLVNGAALA DSYPAGPVVV TLKAKAGYKL GESATTEWKF TVPATPEFAS DTGKLQIPTT | 1080 |
| VGVSYDAQNQ TMAPGKSVTV TATASDGYSF AATRAVKWTF TADQKDDSKP GTDPEKPGTD | 1140 |
| PEKPGTVPSD NPTRPDNGTV ADGKQKNPNK PATSVKDKQG SLVTTGSLAG IAAIAALTLG | 1200 |
| AAGMAIVRRR LKEQ | 1214 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH136 | 40 | 659 | 4.2e-135 | 0.9918533604887984 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13229 | Beta_helix | 3.69e-04 | 280 | 363 | 9 | 92 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADP35567.1 | 1.03e-168 | 40 | 1036 | 68 | 974 |
| ADO52688.1 | 7.95e-167 | 40 | 957 | 34 | 890 |
| QTQ17888.1 | 1.98e-165 | 40 | 957 | 34 | 892 |
| EFR49825.1 | 7.91e-164 | 40 | 957 | 21 | 877 |
| VEG19581.1 | 9.24e-164 | 40 | 957 | 34 | 892 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7V6M_A | 1.20e-165 | 35 | 658 | 4 | 574 | ChainA, Fibronectin type III domain-containing protein [Tyzzerella nexilis] |
| 5GQC_A | 4.41e-130 | 40 | 659 | 18 | 595 | Crystalstructure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_C Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_D Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_E Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_F Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_G Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_H Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQF_A Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, lacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQF_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, lacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQG_A Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, galacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQG_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, galacto-N-biose complex [Bifidobacterium longum subsp. longum] |
| 7V6I_A | 1.40e-127 | 40 | 659 | 14 | 607 | ChainA, Lacto-N-biosidase [Bifidobacterium saguini DSM 23967] |
| 6KQT_A | 7.27e-94 | 42 | 518 | 247 | 662 | CrystalStructure of GH136 lacto-N-biosidase from Eubacterium ramulus - native protein [Eubacterium ramulus ATCC 29099] |
| 6KQS_A | 5.68e-90 | 42 | 518 | 247 | 662 | CrystalStructure of GH136 lacto-N-biosidase from Eubacterium ramulus - selenomethionine derivative [Eubacterium ramulus ATCC 29099] |
SignalP and Lipop Annotations help
This protein is predicted as SP
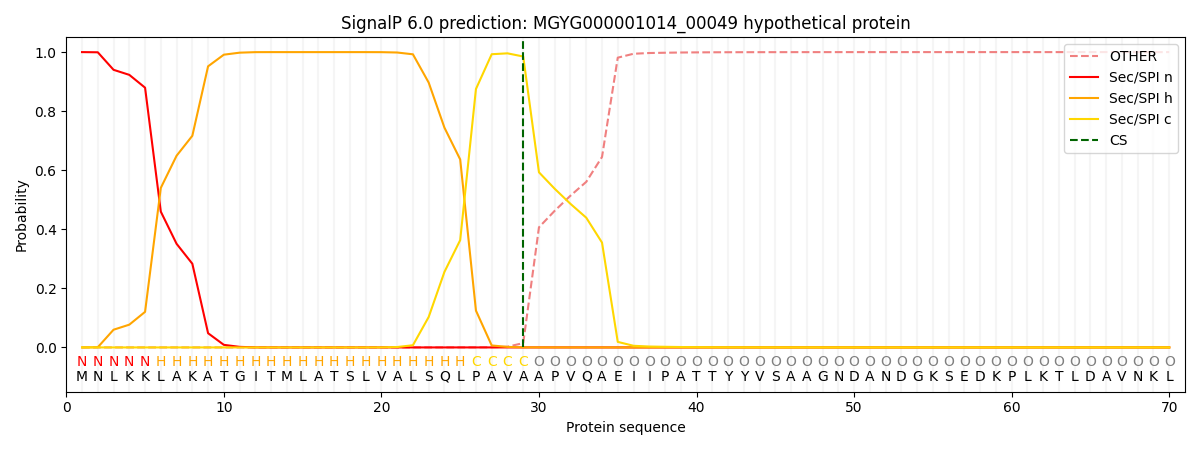
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000279 | 0.999026 | 0.000162 | 0.000189 | 0.000159 | 0.000147 |