You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001210_00067
You are here: Home > Sequence: MGYG000001210_00067
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
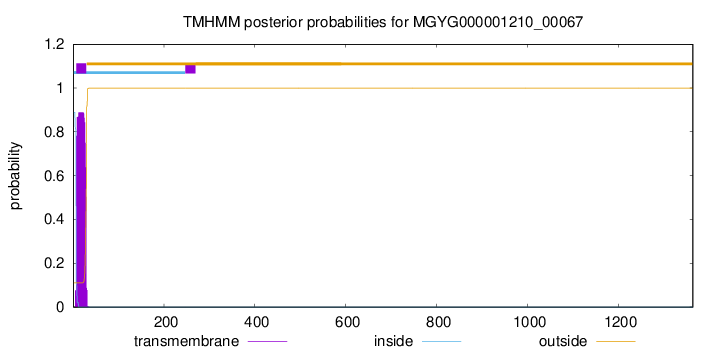
TMHMM annotations
Basic Information help
| Species | Mediterraneibacter sp900553885 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Mediterraneibacter; Mediterraneibacter sp900553885 | |||||||||||
| CAZyme ID | MGYG000001210_00067 | |||||||||||
| CAZy Family | GH146 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 9824; End: 13918 Strand: - | |||||||||||
Full Sequence Download help
| MKKKKLSFIA ALSVFSMTVT SVLPAMAAPT ANDAGKAGDA LENQVLDLEF EGDLKDGTDT | 60 |
| IADISISGDG YEYVDGVNGG KALSLTGNTY VNLGKDQRLQ PENLTLSFWI NPNETITGEQ | 120 |
| IISWNKQEYY TDGWYLTSEN DNTPLALSIG EGANGGQPYK VSVKGNRAEF FPAGEWTHIV | 180 |
| VTYDSETKDV AFYRNGVAQS VNVDYAVGEG RDGLLGSDPD MEKSIGYNGP AYKGAYMNAA | 240 |
| LDHYELFNDT ATAEQALALY EENKETETSG LKNQVLDLEF EDNLTDGTGI NTDIKFSGSG | 300 |
| YEYVDGANGG KALKLTGNTY IDLGRNTELQ PKNLTLSFWI NPEKTMTGEG IISWNKNEYY | 360 |
| TDGWYLSSEG DNVPLALSIG EGKNAGQPYK VSIKGNRAEF FPAGEWTHIV VTYNSASKEV | 420 |
| VFYRNGIEQT STIDYAVNSD RDGVLGSDAS MQKSIGYNGP AYNGSYLKAA LDHYELFNDT | 480 |
| ATKEEAMALY EENGGEIDKA QIAKDDLDAL NIPAETSRNL ALPSVGEQGS VITWEVTAGT | 540 |
| AMDNNGVITR PEIGEPDAKV TLTATAVFMD GEAATKTFDV TVLAKKPMDL SDTSLMDDVT | 600 |
| LADDYMVNAF TKEEEYLLSL NSEKFLYEFY NVAGLEPTTE EGYGGWERTG ASNFRGHTFG | 660 |
| HYMSALSQAY LSEDDADVKA QLMEQIEDAV NGLKKCQDAY AAMYPQSAGY ISAFPEGVLA | 720 |
| RVDGGTSPTP DDGTVLVPYY NLHKVVAGLI DVAKNVDDTE IKNTAIEVAE AFGEYLYNRC | 780 |
| TNLPNKQQLL WIEYGGMNEA LYELYRITGN DHIKAAAECF DEVSLFQQLA AGQDVLSGKH | 840 |
| ANTTIPKFTG AVKRYSVLTE NKEYYDRLTQ EEKDNLDMYL TAAENFWDIV VNDHTYITGG | 900 |
| NSQSEHFHNA GELCYDATKG SYDGALTCET CNTYNMLKLT KALYDQTQDP KYLDFFERTF | 960 |
| TNAILASQNP ETGTTMYFQP MAAGYNKVYN RPYDEFWCCT GTGMENFSKL GDYIYQIKGN | 1020 |
| TLYVNMFYSS TVEDGGLGIS LTQDANIPNE DTVIYTVNDI ADGTSVALRQ PEWLAGDAVI | 1080 |
| KVNGEVKDVT AENGFFVLEG LKAGDEISYT MPMQVEIEST ADNENFVAFK YGPVVLAAEL | 1140 |
| GSKDIDASQA NGILVRVGTE DTSAKDTITV QNMSVEEWKS NITENFVRIE DSEDGRVQFQ | 1200 |
| LKNTDSEELI YSPYYMQHEQ RYGIYMNLEE PDSQASQDRI LAEKETLREQ EVSVDYLDSF | 1260 |
| DNNNSEFAKN LQTGGDTSTG TFNGRTFRHA NAGGWWSYDL AVDPEADHNY LGCTWYSGDQ | 1320 |
| GRSFEIYVND SLLKTVQINN AAGTNVFYED LYDITDYIKV GLRK | 1364 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH146 | 598 | 1137 | 6.3e-168 | 0.9980119284294234 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07944 | Glyco_hydro_127 | 6.52e-125 | 598 | 1137 | 1 | 503 | Beta-L-arabinofuranosidase, GH127. One member of this family, from Bidobacterium longicum, UniProtKB:E8MGH8, has been characterized as an unusual beta-L-arabinofuranosidase enzyme, EC:3.2.1.185. It rleases l-arabinose from the l-arabinofuranose (Araf)-beta1,2-Araf disaccharide and also transglycosylates 1-alkanols with retention of the anomeric configuration. Terminal beta-l-arabinofuranosyl residues have been found in arabinogalactan proteins from a mumber of different plantt species. beta-l-Arabinofuranosyl linkages with 1-4 arabinofuranosides are also found in the sugar chains of extensin and solanaceous lectins, hydroxyproline (Hyp)2-rich glycoproteins that are widely observed in plant cell wall fractions. The critical residue for catalytic activity is Glu-338, in a ET/SCAS sequence context. |
| COG3533 | COG3533 | 9.11e-82 | 599 | 1141 | 12 | 505 | Uncharacterized conserved protein, DUF1680 family [Function unknown]. |
| pfam13385 | Laminin_G_3 | 6.12e-15 | 85 | 255 | 1 | 149 | Concanavalin A-like lectin/glucanases superfamily. This domain belongs to the Concanavalin A-like lectin/glucanases superfamily. |
| pfam13385 | Laminin_G_3 | 7.84e-13 | 315 | 485 | 1 | 149 | Concanavalin A-like lectin/glucanases superfamily. This domain belongs to the Concanavalin A-like lectin/glucanases superfamily. |
| cd00152 | PTX | 7.73e-06 | 80 | 220 | 6 | 139 | Pentraxins are plasma proteins characterized by their pentameric discoid assembly and their Ca2+ dependent ligand binding, such as Serum amyloid P component (SAP) and C-reactive Protein (CRP), which are cytokine-inducible acute-phase proteins implicated in innate immunity. CRP binds to ligands containing phosphocholine, SAP binds to amyloid fibrils, DNA, chromatin, fibronectin, C4-binding proteins and glycosaminoglycans. "Long" pentraxins have N-terminal extensions to the common pentraxin domain; one group, the neuronal pentraxins, may be involved in synapse formation and remodeling, and they may also be able to form heteromultimers. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDP74899.1 | 0.0 | 275 | 1361 | 47 | 1144 |
| QUB98427.1 | 0.0 | 275 | 1361 | 47 | 1144 |
| QJW37681.1 | 0.0 | 271 | 1361 | 43 | 1144 |
| ARK06383.1 | 0.0 | 275 | 1361 | 47 | 1144 |
| ARU52215.1 | 0.0 | 275 | 1361 | 47 | 1144 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6YQH_AAA | 2.91e-81 | 596 | 1361 | 30 | 778 | ChainAAA, Acetyl-CoA carboxylase, biotin carboxylase [Bacteroides thetaiotaomicron VPI-5482] |
| 5OPJ_A | 3.23e-78 | 596 | 1361 | 30 | 778 | Beta-L-arabinofuranosidase[Bacteroides thetaiotaomicron] |
| 6EX6_A | 1.30e-16 | 739 | 1139 | 142 | 547 | TheGH127, Beta-arabinofuranosidase, BT3674 [Bacteroides thetaiotaomicron VPI-5482],6EX6_B The GH127, Beta-arabinofuranosidase, BT3674 [Bacteroides thetaiotaomicron VPI-5482] |
| 5HO0_A | 8.40e-07 | 413 | 603 | 470 | 657 | Crystalstructure of AbnA (closed conformation), a GH43 extracellular arabinanase from Geobacillus stearothermophilus [Geobacillus stearothermophilus],5HO2_A Crystal structure of AbnA (open conformation), a GH43 extracellular arabinanase from Geobacillus stearothermophilus [Geobacillus stearothermophilus],5HOF_A Crystal structure of AbnA, a GH43 extracellular arabinanase from Geobacillus stearothermophilus, in complex with arabinopentaose [Geobacillus stearothermophilus],5HP6_A Structure of AbnA, a GH43 extracellular arabinanase from Geobacillus stearothermophilus (a new conformational state) [Geobacillus stearothermophilus] |
| 5HO9_A | 1.29e-06 | 413 | 583 | 470 | 637 | Structureof truncated AbnA (domains 1-3), a GH43 arabinanase from Geobacilllus stearothermophilus, in complex with arabinooctaose [Geobacillus stearothermophilus],5HO9_B Structure of truncated AbnA (domains 1-3), a GH43 arabinanase from Geobacilllus stearothermophilus, in complex with arabinooctaose [Geobacillus stearothermophilus] |
SignalP and Lipop Annotations help
This protein is predicted as SP
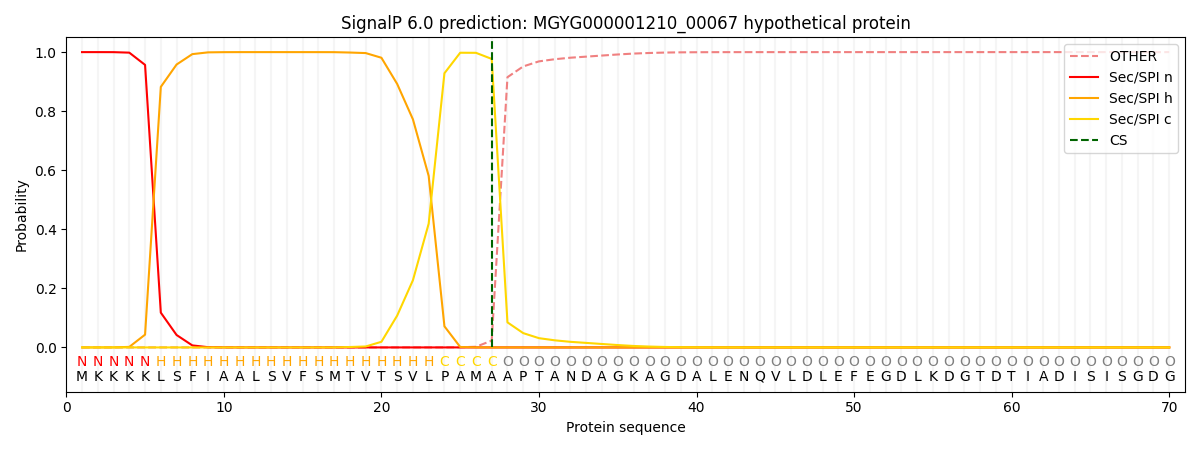
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000266 | 0.998845 | 0.000217 | 0.000232 | 0.000202 | 0.000183 |