You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000029_00068
You are here: Home > Sequence: MGYG000000029_00068
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides finegoldii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides finegoldii | |||||||||||
| CAZyme ID | MGYG000000029_00068 | |||||||||||
| CAZy Family | GH147 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 95776; End: 98385 Strand: - | |||||||||||
Full Sequence Download help
| MKRLFILTSI CLLFAFVNIQ GKSSSTPFIY IDGNGVIRWS DTRQEASFFG VNYTLPFAHA | 60 |
| YRALGYLGLD RKAAIDKDVY HITRLGLNAY RIHLWDVELT DEQGNLLENE HLDLMDYLIA | 120 |
| KLKERNIHIV ITAQTNFGNG YPERNIQTGG FSYKYDKCDV HSHPEAITAQ ETYLHNLVKH | 180 |
| INPYTGIAYK DDPSIVGFEI NNEPCHSGTK EEVKAYINRM LEAIHRTGNR KPVFYNVSHN | 240 |
| EYVVEAYYET AIQGTTYQWY PIGLVSGQTQ QGNFLPYIDR YDISFADKVK GFHKKARLIY | 300 |
| EFDPADIMYS YMYPAMARTF RMAGFQWVTQ FAYDPMDIAY ANTEYQTHFL NLAYTPHKAI | 360 |
| SMKIAAEAAR NLRRGESYGS YPQDTLFGDG FRVSYTENLS ELNNGRKYYY SNNTHSLPQN | 420 |
| ASQLMSIAGC GSSPVVHYEG TGAYFIDCLE AGVWRLEVMP DAVTVSDPFA KPSLAKEVVS | 480 |
| IIYGCWDMAL QIPDLGESFT LTALNKENQR KEETVKDGVI RNLQPGVYLL KRKNCTPKQN | 540 |
| WTTDSRWNSI RLGEFAAPSP HTTDYKVIHN PAQVTEADRD LTILAQVVGN TSPDSVIIYT | 600 |
| DKVSFWNDHN PSIKMKRISG YSYQATIPAA ELQEGCFKYN IIVCRGDSTR TYPAGNSVKG | 660 |
| FSSGIKGNPL DWDYIADSYW TTRIVAPDSP IQLLSMTDTH SGIEAYTLPE WNDLKRTLID | 720 |
| FSPIEKPLLR FAFTSKGEKT QHFLRKSIQD VLKERKGKLK SCTTLCIRVN RHKNLPEGFY | 780 |
| AGFITSDGYT YKTICSTPSP EGIIRIPLKK LHQADTALLP IAYPTFLKQY FHPETDIPFQ | 840 |
| MEKAEKLELS LLGKDKTESI IELGEIWLE | 869 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH147 | 20 | 850 | 0 | 0.991421568627451 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIU95131.1 | 0.0 | 1 | 869 | 1 | 869 |
| QGT73999.1 | 0.0 | 1 | 869 | 1 | 869 |
| QRQ56251.1 | 0.0 | 1 | 869 | 1 | 869 |
| SCV08893.1 | 0.0 | 1 | 869 | 1 | 869 |
| ALJ49469.1 | 0.0 | 1 | 869 | 1 | 869 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4QP0_A | 2.58e-07 | 85 | 204 | 59 | 176 | CrystalStructure Analysis of the Endo-1,4-beta-mannanase from Rhizomucor miehei [Rhizomucor miehei] |
SignalP and Lipop Annotations help
This protein is predicted as SP
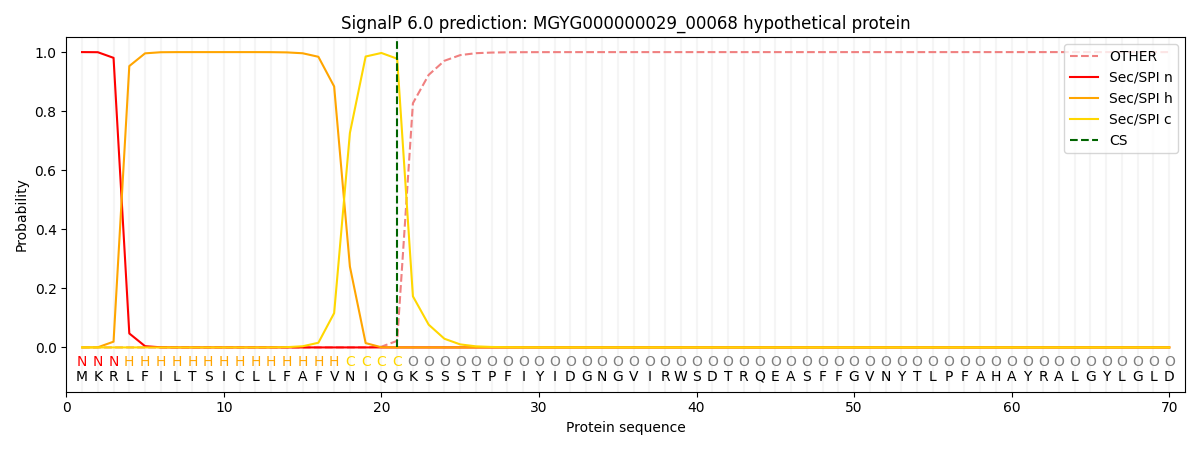
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000298 | 0.998984 | 0.000229 | 0.000165 | 0.000155 | 0.000146 |
