You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001546_00144
You are here: Home > Sequence: MGYG000001546_00144
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes provencensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; Alistipes provencensis | |||||||||||
| CAZyme ID | MGYG000001546_00144 | |||||||||||
| CAZy Family | GH163 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 145449; End: 147599 Strand: - | |||||||||||
Full Sequence Download help
| MCSCISRFKR LALVASAALC CSFEGVGCAS AGVSPAACDG YAVLPADDLS REDLRWSEYL | 60 |
| YDHLWRRAGG GERTVGREEG AKLFTLTVGV DTLLRADFSV KRFRRGVRLT ARDGRRMLWL | 120 |
| QYQLMKSLAG EDPRIEASDL PPATVGLRDT SGRFAFGFRG LYTPAAHDAD HAGILAADDA | 180 |
| ETGWGLWGHQ LERVLADGPE EVYALIDGER YDGQFCFSAD ETYRRIEEYI TDNFGEGDTA | 240 |
| ALRFVVAPTD DDAICSCAAC TAVGNTAKSA TPAVVKLLTR LAGRFPHHTF FTLGYLSTLE | 300 |
| PPPVAMPANA GVIVSAMDLP FGADAFKSAR GKKFAGRLDR WKNVAGEVYI WDYIQNFDDY | 360 |
| LTPFPVLELT ARRLRFYRDR GVGGVFLNGS GCDYVPFDDM RTYVLSALML DPGQSLEALM | 420 |
| RRYFAENYPR SGELLAGFCA DAERRAAASK RPLNLYGSIR SAEEEWLDAG DFVRFYEALE | 480 |
| AVLPTTKDAE RRSLHRLLTA LSYTRLEIAR NHAAEACGFA VNKDGRLRVN PSVERWLAAL | 540 |
| DTHADFPEME HINESGLLPG DYLAQWRGRL LPGTGVRNLL LGVKPEIVSK PDEEYKDPGV | 600 |
| LTDGVRGLPL GYHYGWHISS GDLEVAFPAG YGADARRFEM SFLELPRHRL RAPRAVEIYK | 660 |
| DGALYRSFVP APDAEGRVFT ASGPVDLAGA KRISVRAVRP DGKRVQLAAD EIYLIP | 716 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH163 | 199 | 435 | 2.5e-62 | 0.9402390438247012 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam16126 | DUF4838 | 1.53e-58 | 187 | 438 | 1 | 262 | Domain of unknown function (DUF4838). This family consists of several uncharacterized proteins found in various Bacteroides and Chloroflexus species. The function of this family is unknown. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT74209.1 | 2.39e-202 | 39 | 716 | 41 | 719 |
| QIU93722.1 | 3.75e-189 | 39 | 716 | 41 | 717 |
| QUT26575.1 | 4.78e-187 | 39 | 716 | 41 | 717 |
| QGT72808.1 | 1.91e-186 | 39 | 716 | 41 | 717 |
| QRM99284.1 | 1.91e-186 | 39 | 716 | 41 | 717 |
SignalP and Lipop Annotations help
This protein is predicted as SP
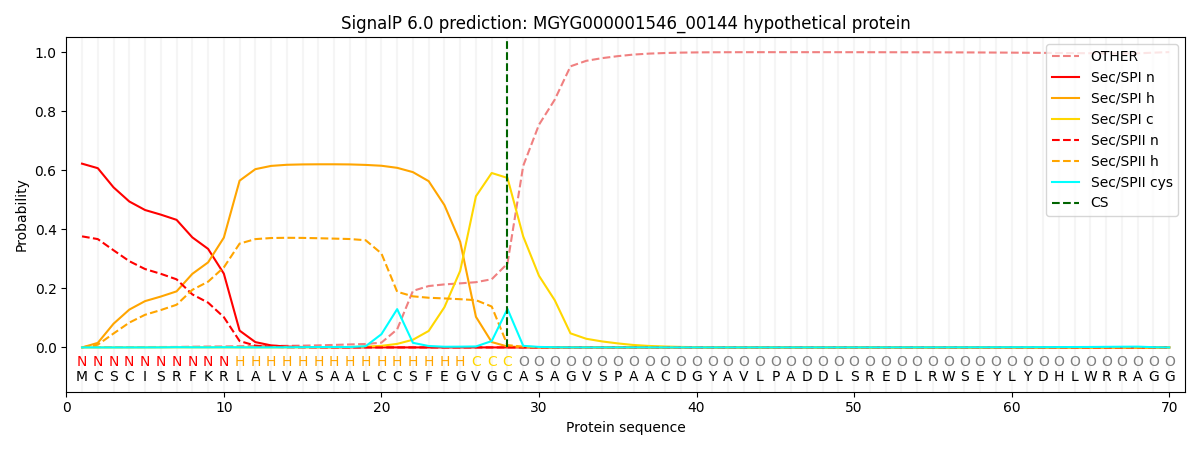
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002638 | 0.608149 | 0.387628 | 0.000837 | 0.000436 | 0.000282 |
