You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003237_00049
You are here: Home > Sequence: MGYG000003237_00049
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Victivallis sp002998355 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; Victivallaceae; Victivallis; Victivallis sp002998355 | |||||||||||
| CAZyme ID | MGYG000003237_00049 | |||||||||||
| CAZy Family | GH167 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 410; End: 4492 Strand: - | |||||||||||
Full Sequence Download help
| MRQRKRILLA GCAAAAALTA SADSNLVRNG SFERAGGDSV PLEWNFSRNG DVPVTAFSTT | 60 |
| PGAEGEKCLR IVNRQTGKKP NRFGLLSQTV SLRPDTDYLF SYKIRGPKET NANWAFGSKW | 120 |
| LIRRPVAGAS GGGWTQHQFV LRVPADRMEG ENLCGVRLIT EGPAEFDIDD VRLVPMGDNL | 180 |
| LLNGSFDGIP GQLPPGWSYR ISGGADVTGA VDAASALTGK TSFKFVNTTP KNANVYGVLT | 240 |
| QIVRLQPEID YVLKVHARGE GQGITLAVGS KWAHRLGLQP LTDKWRTYEM PFRLAKDEVG | 300 |
| KDGSTPVVII SEDVAPGVWI DDISVVPKTA PNLGSALWQK NRVYAVDRLP GKFDELKTVP | 360 |
| AGLPVFRLPL SSENAADGKM PDAKNFSADV ALGYDDAGLI LLARVNDDVS LPGAGSDMWQ | 420 |
| RDSIQIRIDR AAAFAAMSSD TDFEAGFSVG RDGAVQSWCW DGGAQAGKPL PAELAETRVF | 480 |
| RTEQGYFLAA RLRWELLGDV KKSGKFGFTI VINDSDSATH RTAYFLTPGL HDKKYATQYI | 540 |
| QALLDTGSPG VLAKLPSTPS AQILEGRLLL SRAPENAVLT AELSDAAGKK FLRSIADIRG | 600 |
| VKPGELVLAP FSLTLDGLAK GNYTVDFKLN GKSVGTAQAV KADLYEQQNE LLTELCARLE | 660 |
| RLKKEFASVY GDRPYAVCVS VPIAVLDRHL PLLRKRLKNA SGDGEKLFYA EQAAMTARET | 720 |
| ADMLDLLEEN LNTLRGGGRL PAAWKYQSGT ITLENGWPVA DAVCEDGRRE RRPVIFTGFG | 780 |
| HFGDIDRDMT VFPGMGVNVV QIELGPSRLF PREGKNREFE PDYSIVNSRI LPMLEKAWKN | 840 |
| NNTIALLISP HYHPDWLLKK YPELAAPSGF LKYEVNQPKA REMVREYIAA LFGKLKESPY | 900 |
| FGAIHSICLT NEPVYTACRP DNPFSRQEFK KYMEKKYGSV AEFNKVAGTG FADCGAMLDA | 960 |
| VTGNDPAAKY EFYTFAREAF ADWHRLLAEE VKKAVPGMPV HTKIMVFSSP FEYASGVDPE | 1020 |
| LMGDFSEYNG NDNYFYRRGR WIADWDVTAM THEIQMSAKP MSIANTENHI IPDGETRPVS | 1080 |
| NDHIYTANFQ QFITGASTLA TWVWADYQYD FAKQNPSHSF IGNIFLRPGN IAAHCLSGLD | 1140 |
| GLRLAPEIRK FTDDRPEVAL LYSPTATILN SGSYRAETDA LYTALCFTGY RPRFLTERQL | 1200 |
| ARGEFGNSRL LYVAGAKNVS RAARLGMKKF TENGGRIALT PGSLAQDEFG RPAAPDFPTE | 1260 |
| PAAPENAAAL TAQIHRSGVA PLPAAVKVAH EDGNSGIFFR MVPDGKGAWL VNLVNYNFEP | 1320 |
| RRIQLEGAGK WFDLIREADF QPEFELAPLK PQLLRFTPAE | 1360 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH167 | 709 | 1355 | 9e-138 | 0.9093444909344491 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd09621 | CBM9_like_5 | 1.07e-18 | 384 | 544 | 26 | 188 | DOMON-like type 9 carbohydrate binding module. Family 9 carbohydrate-binding modules (CBM9) play a role in the microbial degradation of cellulose and hemicellulose (materials found in plants). The domain has previously been called cellulose-binding domain. The polysaccharide binding sites of CBMs with available 3D structure have been found to be either flat surfaces with interactions formed by predominantly aromatic residues (tryptophan and tyrosine), or extended shallow grooves. CBM9 domains found in this uncharacterized heterogeneous subfamily are often located at the C-terminus of longer proteins and may co-occur with various other functional domains such as glycosyl hydrolases. The CBM9 module in these architectures may be involved in binding to carbohydrates. |
| cd09619 | CBM9_like_4 | 2.51e-13 | 368 | 516 | 18 | 161 | DOMON-like type 9 carbohydrate binding module. Family 9 carbohydrate-binding modules (CBM9) play a role in the microbial degradation of cellulose and hemicellulose (materials found in plants). The domain has previously been called cellulose-binding domain. The polysaccharide binding sites of CBMs with available 3D structure have been found to be either flat surfaces with interactions formed by predominantly aromatic residues (tryptophan and tyrosine), or extended shallow grooves. CBM9 domains found in this uncharacterized heterogeneous subfamily are often located at the C-terminus of longer proteins and may co-occur with various other domains. |
| COG1874 | GanA | 2.21e-11 | 784 | 1059 | 31 | 303 | Beta-galactosidase GanA [Carbohydrate transport and metabolism]. |
| pfam02449 | Glyco_hydro_42 | 3.77e-10 | 785 | 1059 | 12 | 288 | Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues. |
| pfam06452 | CBM9_1 | 7.68e-10 | 351 | 522 | 3 | 157 | Carbohydrate family 9 binding domain-like. CBM9_1 is a C-terminal domain on bacterial xylanase proteins, and it is tandemly repeated in a number of family-members. The CBM9 module binds to amorphous and crystalline cellulose and a range of soluble di- and monosaccharides as well as to cello- and xylo- oligomers of different degrees of polymerization. Comparison of the glucose and cellobiose complexes during crystallisation reveals surprising differences in binding of these two substrates by CBM9-2. Cellobiose was found to bind in a distinct orientation from glucose, while still maintaining optimal stacking and electrostatic interactions with the reducing end sugar. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM43323.1 | 0.0 | 1 | 1360 | 1 | 1360 |
| AVM46829.1 | 8.46e-236 | 173 | 1359 | 16 | 1183 |
| AVM43886.1 | 5.83e-135 | 551 | 1354 | 196 | 996 |
| AHF94331.1 | 1.86e-95 | 13 | 1355 | 16 | 1387 |
| QHW30800.1 | 4.13e-89 | 361 | 1252 | 498 | 1367 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3TTS_A | 7.62e-07 | 785 | 1059 | 25 | 298 | ChainA, Beta-galactosidase [Niallia circulans subsp. alkalophilus],3TTS_B Chain B, Beta-galactosidase [Niallia circulans subsp. alkalophilus],3TTS_C Chain C, Beta-galactosidase [Niallia circulans subsp. alkalophilus],3TTS_D Chain D, Beta-galactosidase [Niallia circulans subsp. alkalophilus],3TTS_E Chain E, Beta-galactosidase [Niallia circulans subsp. alkalophilus],3TTS_F Chain F, Beta-galactosidase [Niallia circulans subsp. alkalophilus],3TTY_A Chain A, Beta-galactosidase [Niallia circulans subsp. alkalophilus],3TTY_B Chain B, Beta-galactosidase [Niallia circulans subsp. alkalophilus],3TTY_C Chain C, Beta-galactosidase [Niallia circulans subsp. alkalophilus],3TTY_D Chain D, Beta-galactosidase [Niallia circulans subsp. alkalophilus],3TTY_E Chain E, Beta-galactosidase [Niallia circulans subsp. alkalophilus],3TTY_F Chain F, Beta-galactosidase [Niallia circulans subsp. alkalophilus] |
SignalP and Lipop Annotations help
This protein is predicted as SP
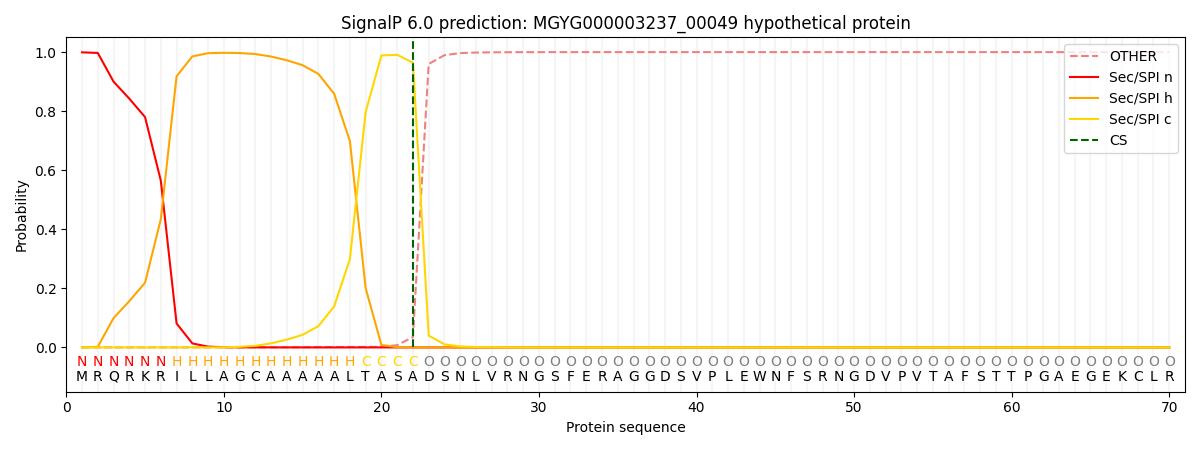
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001172 | 0.996603 | 0.000357 | 0.001296 | 0.000315 | 0.000236 |
