You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001159_00252
You are here: Home > Sequence: MGYG000001159_00252
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
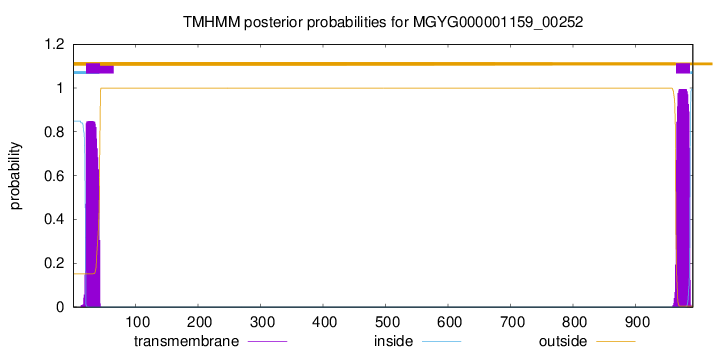
TMHMM annotations
Basic Information help
| Species | Bifidobacterium pullorum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Bifidobacteriaceae; Bifidobacterium; Bifidobacterium pullorum | |||||||||||
| CAZyme ID | MGYG000001159_00252 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 123613; End: 126591 Strand: - | |||||||||||
Full Sequence Download help
| MGASHVFPQG GTTVKRRIAA IAAGIATAAM ALGGVVAPPA TAYADEPAAA DVSSVTNPNN | 60 |
| RHFMVYYRAW RDVTMKGVNT DLPDENWISM YDIPYGVDVV NVFSYVPAGQ EAAAQPFYDK | 120 |
| LKAEYAPYLH SRGIKLVRGI NYEQVTVEGF RDYMADLGKT ADEATEADYD AYALEIIDEY | 180 |
| MTSVGLDGLD IDMETYPTDA DVAISDNVIR ALSKHIGPKS DDPDGTMFLY DTNGSNTKPF | 240 |
| ANVADCFDYV AYQQYGSTST RTAGAVADYS PYIGADKFVP GLAFPEEGDH NRWYDATEPY | 300 |
| TDSHIYDIAS YVRDNGLGGM FLYALDRDGR TYEAEDWSHI VPSNLLWTKT AIAESQDMTM | 360 |
| EQAKAAANHY IERMSLTEEG AAGVALNAED ALEAVEQATN LYEVNKAVLG GDYDEGFSNT | 420 |
| YDPTLEVGLL GVDTTELMDR ISAADNILAG DVMSDDVKSA VRQSRDAAVE GLTGRTYTAD | 480 |
| EVAMWTADLQ ADIEAAMASL TGVESSDRHF MVYYRAWRDV TMKGVNTDLP DENWISMYDI | 540 |
| PYGVDVVNVF SYVPAGQEAA AQPYYDKLKS DYAPYLHSRG IKLVRGVDYT GVIVDGFRDF | 600 |
| IEAKGLTEQE ATEADYDEYA LQVIDEYMTS VGLDGLDIDM ETHPDEADVA VSDNVIRALS | 660 |
| KHIGPKSDNP DGTMFLYDTN ASDLAPFRNV ADCFDYVAYQ QYGSNAERTD DAFGDYAPHI | 720 |
| GSEFVPGLTF PEEGDMNNRW YDATEPYEES NFYQVASYVN EHKLGGMFVY ALDRDGRDYE | 780 |
| DDLTRIVPSN LLWTKTAIAE SQGMPLDQAK AAAKHYIERM SLASAEGVAL NADEAGAAVA | 840 |
| AAANLYEVNK AVLGGDYGEG FSNTYDPTLE AGLLDIDIDE LMSLIEQADK ALEAEDDDTV | 900 |
| RTARDAAMDG LTGKIYTADE VGQWVDALTA ALKGEETPSG PDDTSKPNTD VQKPADEKPL | 960 |
| ASSGSAVAAV LSVAVAAAVA GIGLTVWRRR KV | 992 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd06542 | GH18_EndoS-like | 1.60e-45 | 62 | 336 | 2 | 249 | Endo-beta-N-acetylglucosaminidases are bacterial chitinases that hydrolyze the chitin core of various asparagine (N)-linked glycans and glycoproteins. The endo-beta-N-acetylglucosaminidases have a glycosyl hydrolase family 18 (GH18) catalytic domain. Some members also have an additional C-terminal glycosyl hydrolase family 20 (GH20) domain while others have an N-terminal domain of unknown function (pfam08522). Members of this family include endo-beta-N-acetylglucosaminidase S (EndoS) from Streptococcus pyogenes, EndoF1, EndoF2, EndoF3, and EndoH from Flavobacterium meningosepticum, and EndoE from Enterococcus faecalis. EndoS is a secreted endoglycosidase from Streptococcus pyogenes that specifically hydrolyzes the glycan on human IgG between two core N-acetylglucosamine residues. EndoE is a secreted endoglycosidase, encoded by the ndoE gene in Enterococcus faecalis, that hydrolyzes the glycan on human RNase B. |
| cd06542 | GH18_EndoS-like | 6.31e-44 | 509 | 781 | 2 | 247 | Endo-beta-N-acetylglucosaminidases are bacterial chitinases that hydrolyze the chitin core of various asparagine (N)-linked glycans and glycoproteins. The endo-beta-N-acetylglucosaminidases have a glycosyl hydrolase family 18 (GH18) catalytic domain. Some members also have an additional C-terminal glycosyl hydrolase family 20 (GH20) domain while others have an N-terminal domain of unknown function (pfam08522). Members of this family include endo-beta-N-acetylglucosaminidase S (EndoS) from Streptococcus pyogenes, EndoF1, EndoF2, EndoF3, and EndoH from Flavobacterium meningosepticum, and EndoE from Enterococcus faecalis. EndoS is a secreted endoglycosidase from Streptococcus pyogenes that specifically hydrolyzes the glycan on human IgG between two core N-acetylglucosamine residues. EndoE is a secreted endoglycosidase, encoded by the ndoE gene in Enterococcus faecalis, that hydrolyzes the glycan on human RNase B. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BAJ70116.1 | 2.50e-228 | 491 | 989 | 34 | 543 |
| QKY14302.1 | 2.50e-228 | 491 | 989 | 34 | 543 |
| VEG44861.1 | 2.50e-228 | 491 | 989 | 34 | 543 |
| ACJ53522.1 | 2.50e-228 | 491 | 989 | 34 | 543 |
| QTB92326.1 | 7.07e-228 | 491 | 989 | 34 | 543 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7PUJ_A | 1.54e-151 | 505 | 932 | 8 | 429 | ChainA, Beta-N-acetylhexosaminidase [Enterococcus faecalis],7PUK_A Chain A, Beta-N-acetylhexosaminidase [Enterococcus faecalis],7PUK_C Chain C, Beta-N-acetylhexosaminidase [Enterococcus faecalis] |
| 6KPL_A | 1.89e-31 | 495 | 798 | 16 | 292 | CrystalStructure of endo-beta-N-acetylglucosaminidase from Cordyceps militaris in apo form [Cordyceps militaris CM01],6KPM_A Crystal Structure of endo-beta-N-acetylglucosaminidase from Cordyceps militaris in complex with L-fucose [Cordyceps militaris CM01] |
| 6KPN_A | 2.16e-30 | 495 | 798 | 16 | 292 | CrystalStructure of endo-beta-N-acetylglucosaminidase from Cordyceps militaris D154N/E156Q mutant in complex with fucosyl-N-acetylglucosamine [Cordyceps militaris CM01],6KPO_A Crystal Structure of endo-beta-N-acetylglucosaminidase from Cordyceps militaris D154N/E156Q mutant in complex with fucosyl-N-acetylglucosamine-Asn [Cordyceps militaris CM01] |
| 6E58_A | 5.03e-18 | 66 | 345 | 28 | 319 | Crystalstructure of Streptococcus pyogenes endo-beta-N-acetylglucosaminidase (EndoS2) [Streptococcus pyogenes M49 591],6E58_B Crystal structure of Streptococcus pyogenes endo-beta-N-acetylglucosaminidase (EndoS2) [Streptococcus pyogenes M49 591],6MDS_A Crystal structure of Streptococcus pyogenes endo-beta-N-acetylglucosaminidase (EndoS2) with complex biantennary glycan [Streptococcus pyogenes],6MDS_B Crystal structure of Streptococcus pyogenes endo-beta-N-acetylglucosaminidase (EndoS2) with complex biantennary glycan [Streptococcus pyogenes],6MDV_A Crystal structure of Streptococcus pyogenes endo-beta-N-acetylglucosaminidase (EndoS2) with high-mannose glycan [Streptococcus pyogenes],6MDV_B Crystal structure of Streptococcus pyogenes endo-beta-N-acetylglucosaminidase (EndoS2) with high-mannose glycan [Streptococcus pyogenes] |
| 4NUY_A | 1.21e-12 | 66 | 338 | 20 | 320 | Crystalstructure of EndoS, an endo-beta-N-acetyl-glucosaminidase from Streptococcus pyogenes [Streptococcus pyogenes serotype M1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B7GPC7 | 5.00e-229 | 491 | 989 | 34 | 543 | Endo-beta-N-acetylglucosaminidase OS=Bifidobacterium longum subsp. infantis (strain ATCC 15697 / DSM 20088 / JCM 1222 / NCTC 11817 / S12) OX=391904 GN=Blon_2468 PE=1 SV=1 |
| P36912 | 3.16e-29 | 497 | 775 | 49 | 307 | Endo-beta-N-acetylglucosaminidase F2 OS=Elizabethkingia meningoseptica OX=238 GN=endOF2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
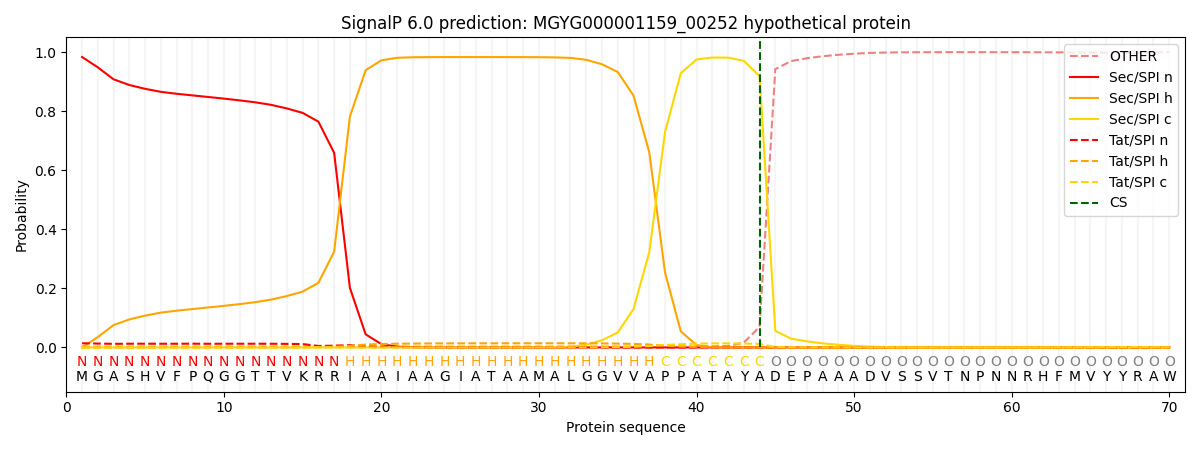
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.004387 | 0.978949 | 0.000457 | 0.015394 | 0.000497 | 0.000295 |