You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000013_00483
You are here: Home > Sequence: MGYG000000013_00483
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides sp902362375 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides sp902362375 | |||||||||||
| CAZyme ID | MGYG000000013_00483 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Evolved beta-galactosidase subunit alpha | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 665502; End: 668930 Strand: - | |||||||||||
Full Sequence Download help
| MLFKKSDTNK NKFESMNKPS FSLLLTGICT GVFSLVTMNA MANSTPRKPY WQDVQVVAVN | 60 |
| KEYPRTAFMT YDNRTDALTG KYENSKYYQL LNGDWKFFFV DSYKKLPENI TDPSVSTASW | 120 |
| NDIKVPGNWE VQGYGVAIYT NHGYEFKPRN PEPPILPEAN PVGVYRRDID IPSDWKERDI | 180 |
| YLHLAGVKSG VYVYINGQEV GYSEDSKNPA EFLINNYVKP GKNVLILKVF RWSTGSYLEC | 240 |
| QDFWRISGIE RDVFLYSQPK AALKDFRVKS TLDDSYKNGI FALNVDLRNR GKEAANLTLV | 300 |
| YELLNAQGKV VAAEEKTAYI PSGEIRAVSF DQNLTNVSTW TSEAPNLYKL LMTVKKNGKV | 360 |
| NEIIPFNVGF RRIEIKPIEQ KAANGKPYVC LFINGQPLKL KGVNIHEHNP ETGHYVTEEL | 420 |
| MRRDFELMKQ HNLNSVRLCH YPQDRRFYEL CDEYGLYAYD EANIESHGMY YNLRKGGTLA | 480 |
| NNPEWLKAHM DRTINMFERN KNYPCVTFWS LGNEAGNGYN FYQTYLWLKE ADKNLMVRPI | 540 |
| NYERAQWEWN SDMYVPQYPG ASWLASVGKN GSDRPIVPSE YAHAMGNSTG NLWGQWQAIY | 600 |
| KYPNLQGGYI WDWVDQGLLQ KDKNGKEYWA YGGDFGVNAP SDGNFLCNGL VNPDRGPHPA | 660 |
| MAEVKYVHQN VGFEAIDAAS GKFRITNRFY FTNLKKYRIH YSVLANGKTI KNGNVSLDIA | 720 |
| PQGSKEFTVS VNGLKVQPGT DYFVNFSVTT TEPEALIPTG YEIAYDQFQL PVQAEKSTYK | 780 |
| ANGPALKTAT QGDEFIVSSS KVNFVFNKNS GLVTSYRVNG TEYFNDGFGI QPNFWRAPTD | 840 |
| NDYGNGAPKR LQVWKQSSKN FCVTDVDMTT KDKVVLLKAT YLLATGNLYV VTYKIYPSGI | 900 |
| VNVKTLFTST DMEAAETEVS EATRTATFTP GSDAARKAAS KLEVPRIGVR FRLPVQMNNV | 960 |
| QYFGRGPGEN YIDRNHGTLV GVYKTTADKM YFNYVRPQEN GHRTDTRWVA LSPDRGNGLA | 1020 |
| IVADSLVGFN ALCNSVEDFD SEEASSRSYQ WHNLTPEEIA NHDENAARNV LRRMHHVNDI | 1080 |
| TPKDFVEVCV DMKQQGVGGY NSWGARPESI HQILANREYS WGFTLVPIHS VNQVNEVTKY | 1140 |
| NY | 1142 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 75 | 979 | 2.9e-207 | 0.976063829787234 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK09525 | lacZ | 0.0 | 51 | 1125 | 17 | 1024 | beta-galactosidase. |
| PRK10340 | ebgA | 0.0 | 51 | 1128 | 4 | 1000 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| COG3250 | LacZ | 2.66e-145 | 91 | 974 | 15 | 800 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| smart01038 | Bgal_small_N | 3.01e-99 | 798 | 1125 | 1 | 272 | Beta galactosidase small chain. This domain comprises the small chain of dimeric beta-galactosidases EC:3.2.1.23. This domain is also found in single chain beta-galactosidase. |
| pfam02836 | Glyco_hydro_2_C | 2.14e-89 | 391 | 673 | 8 | 302 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| SCV08523.1 | 0.0 | 24 | 1142 | 9 | 1126 |
| QRQ55912.1 | 0.0 | 24 | 1142 | 9 | 1126 |
| QNL36573.1 | 0.0 | 24 | 1142 | 9 | 1126 |
| QUT69940.1 | 0.0 | 16 | 1142 | 1 | 1123 |
| ALJ49106.1 | 0.0 | 31 | 1142 | 2 | 1112 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6S6Z_A | 1.43e-233 | 49 | 1125 | 1 | 979 | Structureof beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_B Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_C Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_D Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_E Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_F Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_G Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_H Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8] |
| 6SD0_A | 1.48e-233 | 49 | 1125 | 2 | 980 | Structureof beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_B Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_C Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_D Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8] |
| 3DEC_A | 9.82e-216 | 47 | 1098 | 3 | 969 | ChainA, Beta-galactosidase [Bacteroides thetaiotaomicron VPI-5482] |
| 3BGA_A | 8.88e-213 | 49 | 1128 | 8 | 1001 | Crystalstructure of beta-galactosidase from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482],3BGA_B Crystal structure of beta-galactosidase from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482] |
| 1JZ7_A | 2.57e-149 | 51 | 1018 | 16 | 953 | E.COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH GALACTOSE [Escherichia coli],1JZ7_B E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH GALACTOSE [Escherichia coli],1JZ7_C E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH GALACTOSE [Escherichia coli],1JZ7_D E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH GALACTOSE [Escherichia coli],4TTG_A Beta-galactosidase (E. coli) in the presence of potassium chloride. [Escherichia coli],4TTG_B Beta-galactosidase (E. coli) in the presence of potassium chloride. [Escherichia coli],4TTG_C Beta-galactosidase (E. coli) in the presence of potassium chloride. [Escherichia coli],4TTG_D Beta-galactosidase (E. coli) in the presence of potassium chloride. [Escherichia coli] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q56307 | 8.09e-233 | 49 | 1125 | 2 | 980 | Beta-galactosidase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=lacZ PE=1 SV=2 |
| O52847 | 1.23e-216 | 49 | 1128 | 18 | 1034 | Beta-galactosidase OS=Priestia megaterium (strain DSM 319 / IMG 1521) OX=592022 GN=bgaM PE=3 SV=1 |
| A5F5U6 | 9.67e-180 | 51 | 1125 | 13 | 1016 | Beta-galactosidase OS=Vibrio cholerae serotype O1 (strain ATCC 39541 / Classical Ogawa 395 / O395) OX=345073 GN=lacZ PE=3 SV=2 |
| Q6LL68 | 1.90e-166 | 51 | 1130 | 12 | 1030 | Beta-galactosidase OS=Photobacterium profundum (strain SS9) OX=298386 GN=lacZ PE=3 SV=1 |
| P06864 | 7.75e-165 | 51 | 1128 | 4 | 1000 | Evolved beta-galactosidase subunit alpha OS=Escherichia coli (strain K12) OX=83333 GN=ebgA PE=1 SV=4 |
SignalP and Lipop Annotations help
This protein is predicted as SP
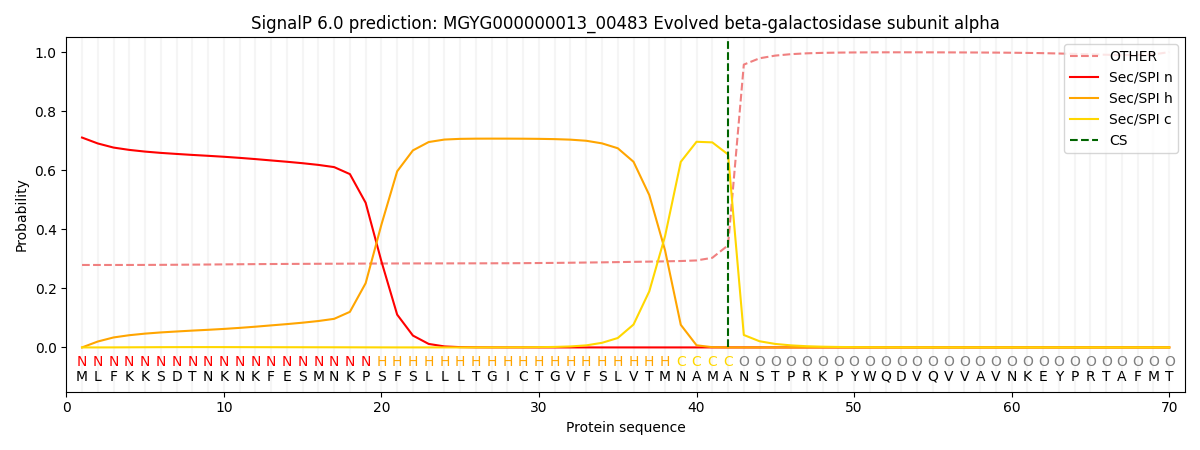
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.301333 | 0.681155 | 0.011575 | 0.003309 | 0.001237 | 0.001389 |