You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000013_01146
You are here: Home > Sequence: MGYG000000013_01146
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides sp902362375 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides sp902362375 | |||||||||||
| CAZyme ID | MGYG000000013_01146 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 121832; End: 125533 Strand: - | |||||||||||
Full Sequence Download help
| MNRISKVFLA CACSVLCMNG QAQSNATWTN DFSNAGETLK VVGRGTCSIA DNVFRSKGAY | 60 |
| AVFGHPEWKN YSFSFKARAP KNAEQVQIWA SFRNYNRFDR YVVGIKGGLQ DDLYLMRTGY | 120 |
| MGTDEFMGVR PLGFHPVPGE WYRVKVEVCG NRIRIFLNDE KQPHIDLVDK NADLVPSGEV | 180 |
| ALGGGWIETE FDDLTVTPMA DDALKNVKVA EYQKKATPAE KENKRCQERA SYTSAKLAAL | 240 |
| TGSRTDITLD GNWLFMPDYQ LDNKDKAVSV QTNDQDWHIM SVPNFWTPIR IWLHGETMPS | 300 |
| PTGAQPKGVS DTYYQQETDR CENYTFDYRR VKYAWYRQWL ELPANVEGKN MTLTFDAVSK | 360 |
| IAEIYINGTL ATSHLGMFGE IQVDGSHLLK PGKNLITVKV TRKMDGSASE SANAIDFFYS | 420 |
| SVRESEQEDA KVEVNKDALL KEIPHGFYGD EPAGIWQPVK LTITDPAKVE DVFIKPTLNG | 480 |
| ATFDVTLKNH GSKKKQFDLY TDIIDKETGA VLYSGLSIRK LNLNADEERM ETYTISDLKP | 540 |
| RLWTPQHPNL YDFKFRLVAD KGTELDCLTE TSGFRTFEVK DGLFYLNGNK YWLRGGNHIP | 600 |
| FALAPNDENL ANTFMQLMKA GNIDVTRTHT TPWNKRWMTA ADRNGIGVSF EGTWSWLMIH | 660 |
| STPIPDQRLI EIWRNEFLGL LKKYRNHPSL LFWTVNNEMK FYDNDSNLER AKEKYRIISD | 720 |
| VVKEMRRIDP TRPICFDSNY QAKGKDKKFG ADFMSSIDDG DIDDMHGYYN WYDYSVFRFF | 780 |
| NGEFQKQFKV TDRPLISQEM STGYPNNETG HPTRSYQLIH QNPYTLIGYE SYDWADPASF | 840 |
| LKVQAFITGE LAETLRRSND QASGIMHFAL MTWFRQTYDY QNIEPYPTYY ALKRALQPVL | 900 |
| VSAELWGRNL YAGEKLPTRI YVVNDREDGT DLQPSLLRWE IQDESGKCLA SGSEKIPAVK | 960 |
| HYARYYAEPD IQLPANLPAD KTKAKLVLKL TENGLPISAN EYELLLTNKE WNVGQVDPNK | 1020 |
| KIVLLDKDNT KMVFDFLNIN YQPISSIKEL LNSKLKADLC VISGLTACTD EEKELIRAYQ | 1080 |
| SKGGKLLFLN SKEAVKAIYP EYITGWIIPT EGDIVIMERN DAPVFNDIDV LELRYFNNNK | 1140 |
| REIPQACTAT LKVHRHKNVT ELAGQMKIHA YIDGGKPEDR IERIESMRGL TMLQIADGKG | 1200 |
| EAMISTMCTE KATTDPVAGK LVVNMINCLT TNK | 1233 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 231 | 1035 | 5.7e-84 | 0.788563829787234 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK10150 | PRK10150 | 1.40e-33 | 240 | 747 | 6 | 459 | beta-D-glucuronidase; Provisional |
| COG3250 | LacZ | 2.34e-29 | 335 | 734 | 66 | 429 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10340 | ebgA | 7.59e-22 | 336 | 746 | 113 | 484 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| pfam00703 | Glyco_hydro_2 | 2.52e-13 | 467 | 575 | 1 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| pfam02836 | Glyco_hydro_2_C | 1.24e-11 | 577 | 799 | 1 | 209 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGT71556.1 | 0.0 | 1 | 1233 | 1 | 1233 |
| QIU96993.1 | 0.0 | 1 | 1233 | 1 | 1232 |
| QDO68995.1 | 0.0 | 1 | 1227 | 1 | 1225 |
| ALJ60854.1 | 0.0 | 1 | 1229 | 1 | 1227 |
| QUT88128.1 | 0.0 | 1 | 1229 | 1 | 1227 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6SD0_A | 3.18e-23 | 247 | 698 | 40 | 441 | Structureof beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_B Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_C Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_D Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8] |
| 6S6Z_A | 3.18e-23 | 247 | 698 | 39 | 440 | Structureof beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_B Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_C Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_D Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_E Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_F Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_G Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_H Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8] |
| 6U7I_A | 1.46e-21 | 304 | 775 | 40 | 476 | Faecalibacteriumprausnitzii Beta-glucuronidase [Faecalibacterium prausnitzii],6U7I_B Faecalibacterium prausnitzii Beta-glucuronidase [Faecalibacterium prausnitzii],6U7I_C Faecalibacterium prausnitzii Beta-glucuronidase [Faecalibacterium prausnitzii],6U7I_D Faecalibacterium prausnitzii Beta-glucuronidase [Faecalibacterium prausnitzii] |
| 6ED1_A | 2.74e-21 | 245 | 772 | 25 | 485 | ChainA, Glycosyl hydrolase family 2, sugar binding domain protein [Phocaeicola dorei],6ED1_B Chain B, Glycosyl hydrolase family 2, sugar binding domain protein [Phocaeicola dorei],6ED1_C Chain C, Glycosyl hydrolase family 2, sugar binding domain protein [Phocaeicola dorei],6ED1_D Chain D, Glycosyl hydrolase family 2, sugar binding domain protein [Phocaeicola dorei] |
| 3GM8_A | 1.51e-19 | 267 | 734 | 24 | 421 | ChainA, Glycoside hydrolase family 2, candidate beta-glycosidase [Phocaeicola vulgatus ATCC 8482] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q56307 | 1.74e-22 | 247 | 698 | 40 | 441 | Beta-galactosidase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=lacZ PE=1 SV=2 |
| P77989 | 2.12e-20 | 327 | 731 | 53 | 412 | Beta-galactosidase OS=Thermoanaerobacter pseudethanolicus (strain ATCC 33223 / 39E) OX=340099 GN=lacZ PE=3 SV=2 |
| O52847 | 1.35e-18 | 248 | 698 | 60 | 481 | Beta-galactosidase OS=Priestia megaterium (strain DSM 319 / IMG 1521) OX=592022 GN=bgaM PE=3 SV=1 |
| Q04F24 | 2.32e-18 | 244 | 708 | 45 | 467 | Beta-galactosidase OS=Oenococcus oeni (strain ATCC BAA-331 / PSU-1) OX=203123 GN=lacZ PE=3 SV=1 |
| B4S2K9 | 5.29e-18 | 336 | 698 | 124 | 460 | Beta-galactosidase OS=Alteromonas mediterranea (strain DSM 17117 / CIP 110805 / LMG 28347 / Deep ecotype) OX=1774373 GN=lacZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
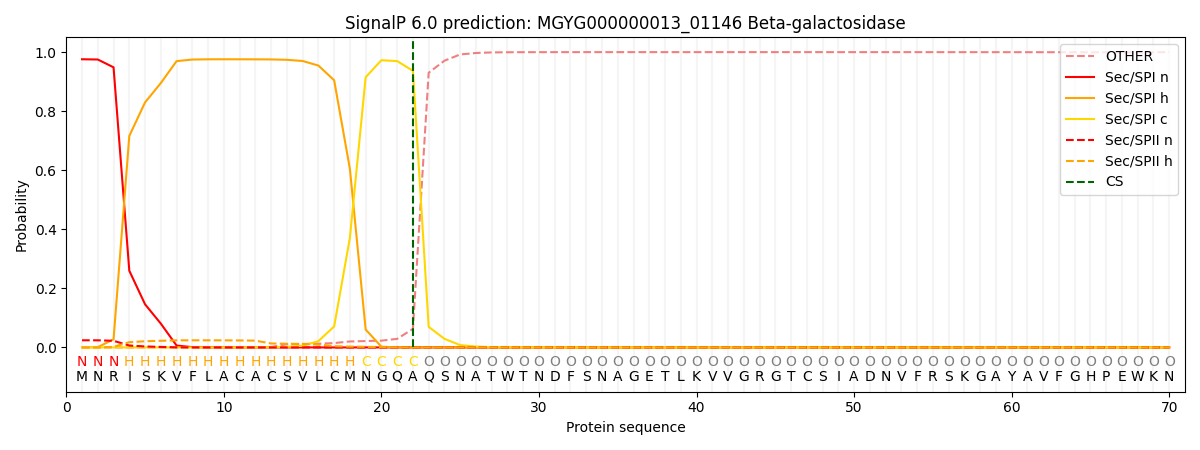
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000448 | 0.969915 | 0.028848 | 0.000284 | 0.000253 | 0.000222 |
