You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000013_01268
You are here: Home > Sequence: MGYG000000013_01268
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides sp902362375 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides sp902362375 | |||||||||||
| CAZyme ID | MGYG000000013_01268 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 308767; End: 311841 Strand: + | |||||||||||
Full Sequence Download help
| MNNLKKRTFL ILSAVLAATT FMLAQQQPLP EWQSQYAVGL NKLAPHTYVW PYANASDIEK | 60 |
| PGGYEQSPFY MSLNGKWKFH WVKNPDNRPK DFYQPSYYTG GWADINVPGN WEPQGYGTAI | 120 |
| YVNETYEFDD KMFNFKKNPP LVPYAENEVG SYRRTFKVPA DWQGRRVVLC CEGVISFYYV | 180 |
| WVNGKLLGYN QGSKTAAEWD ITDVLNEGEN VVALEVYRWS SGAYLECQDM WRLSGIERDV | 240 |
| YLYSTPKQYI ADYKLNASLD KEKYKDGIFG LEVTVEGPSA TTSSIAYTLK DADGKAVLQD | 300 |
| AIRIKSRGLS NFIAFEEKKL PGVKAWSAEH PYLYTLMLEL KDEQGKVTEL TGCEVGFRTS | 360 |
| EIKDGRFCIN GVPVLVKGTN RHEHSQLGRT VSKELMELDI KLMKQHNINT VRNSHYPTHP | 420 |
| YWYQLCDRYG LYMIDEANIE SHGMGYGLAS LAKDSTWLTA HMDRTHRMYE RSKNHPAIVI | 480 |
| WSLGNEAGNG INFERTYDWL KSVEKTRPVQ YERAELNYNT DIYCRMYRSV DDIKAYVNEK | 540 |
| DIYRPFILCE YLHAMGNSCG GLKEYWDVFE NNPMAQGGCV WDWVDQSFRE IDKNGKWYWT | 600 |
| YGGDYGPEGI PSFGNFCCNG LVGANREPHP HLLEVKKVYQ NIKATLANQK NLTIRVKNWY | 660 |
| DFSNLNEYIL NWNVTADNGK ILAEGTKMVD CAPHATVDVT LGAVKFPNTV REAYLNISWT | 720 |
| RREASSMIDK DWEVAYDQFV LAGNKNYTGY RPQKAGETTF TVDKKTGALT SLNLNGKDLL | 780 |
| AAPLTLSLFR PATDNDNRDK NGARLWRDAG LDGLTQKVIS LKESKTSTTA RVEILNAKNQ | 840 |
| KVGTADFVYS LDKNGALKVH TTFQPDTTIV KSIARLGLIF RVANSYDQVS YLGRGDNETY | 900 |
| VDRNQSGKIG VYQTTPERMF HYYVAPQATG NRTDVRWAKL ADTSGEGIFV ESDRAFQFSI | 960 |
| IPFSDVLLEK ARHINELERD GLLTVHLDAE QAGVGTATCG PGVLPQYLVP LKKQSFEFTL | 1020 |
| YPVK | 1024 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 65 | 907 | 3.1e-216 | 0.964095744680851 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK09525 | lacZ | 0.0 | 32 | 1020 | 17 | 1024 | beta-galactosidase. |
| PRK10340 | ebgA | 0.0 | 32 | 1023 | 4 | 1000 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| COG3250 | LacZ | 2.35e-160 | 72 | 910 | 14 | 807 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| pfam02836 | Glyco_hydro_2_C | 7.85e-103 | 361 | 643 | 2 | 301 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| smart01038 | Bgal_small_N | 2.04e-100 | 754 | 1020 | 1 | 272 | Beta galactosidase small chain. This domain comprises the small chain of dimeric beta-galactosidases EC:3.2.1.23. This domain is also found in single chain beta-galactosidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNL38601.1 | 0.0 | 1 | 1024 | 1 | 1024 |
| CBK69498.1 | 0.0 | 3 | 1024 | 6 | 1029 |
| QRN02241.1 | 0.0 | 3 | 1024 | 2 | 1025 |
| QGT74167.1 | 0.0 | 3 | 1024 | 2 | 1024 |
| QRQ57907.1 | 0.0 | 3 | 1024 | 6 | 1028 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3DEC_A | 0.0 | 30 | 1024 | 5 | 998 | ChainA, Beta-galactosidase [Bacteroides thetaiotaomicron VPI-5482] |
| 3BGA_A | 0.0 | 26 | 1024 | 4 | 1002 | Crystalstructure of beta-galactosidase from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482],3BGA_B Crystal structure of beta-galactosidase from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482] |
| 6S6Z_A | 1.55e-227 | 31 | 1019 | 3 | 976 | Structureof beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_B Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_C Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_D Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_E Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_F Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_G Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_H Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8] |
| 6SD0_A | 1.60e-227 | 31 | 1019 | 4 | 977 | Structureof beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_B Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_C Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_D Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8] |
| 1JZ7_A | 7.42e-172 | 31 | 995 | 15 | 994 | E.COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH GALACTOSE [Escherichia coli],1JZ7_B E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH GALACTOSE [Escherichia coli],1JZ7_C E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH GALACTOSE [Escherichia coli],1JZ7_D E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH GALACTOSE [Escherichia coli],4TTG_A Beta-galactosidase (E. coli) in the presence of potassium chloride. [Escherichia coli],4TTG_B Beta-galactosidase (E. coli) in the presence of potassium chloride. [Escherichia coli],4TTG_C Beta-galactosidase (E. coli) in the presence of potassium chloride. [Escherichia coli],4TTG_D Beta-galactosidase (E. coli) in the presence of potassium chloride. [Escherichia coli] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q56307 | 8.76e-227 | 31 | 1019 | 4 | 977 | Beta-galactosidase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=lacZ PE=1 SV=2 |
| O52847 | 5.91e-213 | 30 | 995 | 18 | 1004 | Beta-galactosidase OS=Priestia megaterium (strain DSM 319 / IMG 1521) OX=592022 GN=bgaM PE=3 SV=1 |
| A3FEW8 | 6.51e-192 | 31 | 1019 | 17 | 1024 | Beta-galactosidase OS=Enterobacter agglomerans OX=549 GN=lacZ PE=1 SV=2 |
| A4W7D2 | 2.55e-191 | 31 | 1019 | 17 | 1024 | Beta-galactosidase OS=Enterobacter sp. (strain 638) OX=399742 GN=lacZ PE=3 SV=1 |
| Q47077 | 1.19e-184 | 31 | 1019 | 17 | 1023 | Beta-galactosidase OS=Enterobacter cloacae OX=550 GN=lacZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
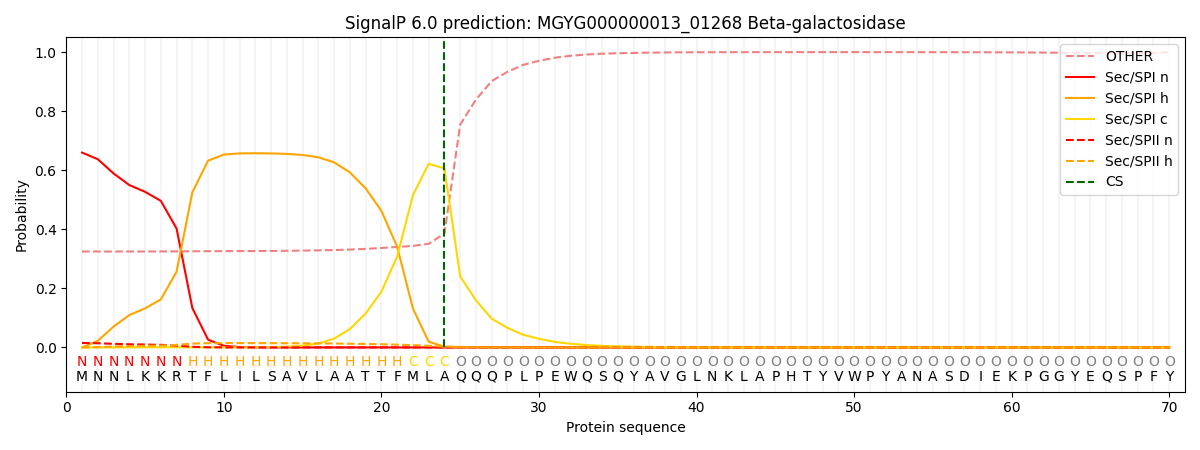
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.343694 | 0.636670 | 0.016733 | 0.001499 | 0.000689 | 0.000690 |
