You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000013_01527
You are here: Home > Sequence: MGYG000000013_01527
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
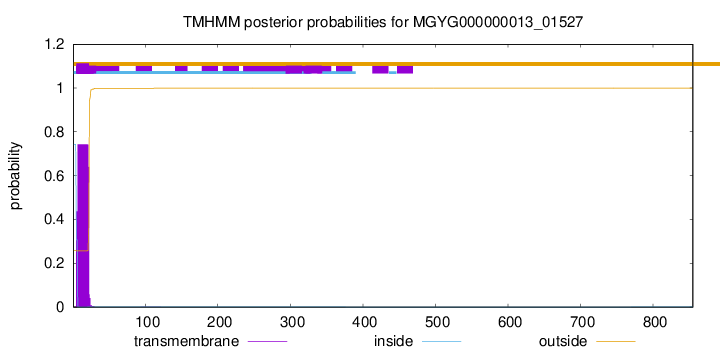
TMHMM annotations
Basic Information help
| Species | Bacteroides sp902362375 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides sp902362375 | |||||||||||
| CAZyme ID | MGYG000000013_01527 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Exo-beta-D-glucosaminidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 602567; End: 605134 Strand: + | |||||||||||
Full Sequence Download help
| MHKKIQYILL LMLFTCLIPV TAKNKSRISL NDEWQFKQSI SQNWLPAQVP GAVHTDLMNN | 60 |
| RMIKDPFYGV NEKSLQWIGE KDWEYKKTFT VDEALLQAPN VQLVFAGLDT YADIYINDYM | 120 |
| VMKCDNMFRT WTLNPLPYLK KGENTIRIYF HSIFKVDMPK YLASPFQLQA WPNNDQSDIM | 180 |
| LSLYARKAGY NYGWDWGPRL ITTGIWRDVY LEYWDDIHID GTQIITRKLE SGKAKMEAVC | 240 |
| TVMSDEDTKA TFTVEYDKKE AVRKQVALTK GMNTVRVDFE VKNPELWWTN GLGNPRMYDF | 300 |
| DIVLDKEGHK VTDRVKTGIR TLEIVREKDE QGQSMFVKLN GKAVFMKGAN YIPLDNFPNR | 360 |
| VPESWYEYII KSAADVNMNM LRIWGGGIYE MDAFYEMCDK YGILVWQDMM FACGMFPADE | 420 |
| HYLNSVAEEV KDNVKRLRNH PCIALWNGNN ENEISYFGWG WKDRYTPEED RIYQSNLHKL | 480 |
| FYDVIPEAIQ AVDETRYYHP TSPVTGYNNI DYNMGDVHFW SVWKGGWLEE YTEAKNIGRF | 540 |
| MSEYGFQSYP EMRTIRRFAS EHDLRLDSEV MLSHQRARND QTRDPNFGNN MMKMYMEKYF | 600 |
| KVPDDFSEFV YMSQYLQAEA VKIGIEAHRR AKPYCMGTLY WQINDCWPVA SWSSIDYYGR | 660 |
| WKALQYYARD AYAEVLVSPY ASGKQVVFKV ISDRLQKMKG ELEITTLTLE GQTVFSKRVS | 720 |
| LELGANGCED VASITTTELY GGQKENEVFT YVTLNENGKT VSSNIYYPVY SNQYAYSKVT | 780 |
| PIITVEKTND GVNLRLRSSA LIRGLYLYVD DDESFFEQNY VTLIPGKEEV VQVKTHLSAA | 840 |
| AFEKQLKYLS VNQVN | 855 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 23 | 800 | 7.4e-105 | 0.7513297872340425 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 3.70e-100 | 26 | 720 | 11 | 679 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| pfam17786 | Mannosidase_ig | 8.00e-15 | 690 | 769 | 7 | 87 | Mannosidase Ig/CBM-like domain. This domain corresponds to domain 4 in the structure of Bacteroides thetaiotaomicron beta-mannosidase, BtMan2A. This domain has an Ig-like fold. |
| pfam00703 | Glyco_hydro_2 | 1.55e-14 | 217 | 320 | 1 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| PRK10340 | ebgA | 7.43e-13 | 85 | 458 | 113 | 453 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| PRK09525 | lacZ | 1.18e-10 | 204 | 451 | 208 | 462 | beta-galactosidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| SCV10456.1 | 0.0 | 1 | 855 | 1 | 855 |
| QRQ57690.1 | 0.0 | 1 | 855 | 1 | 855 |
| ALJ46104.1 | 0.0 | 1 | 855 | 1 | 855 |
| QUT80947.1 | 0.0 | 1 | 855 | 1 | 855 |
| QDM12230.1 | 0.0 | 1 | 855 | 1 | 855 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2VJX_A | 6.33e-194 | 30 | 831 | 11 | 814 | Structuraland biochemical evidence for a boat-like transition state in beta-mannosidases [Bacteroides thetaiotaomicron VPI-5482],2VJX_B Structural and biochemical evidence for a boat-like transition state in beta-mannosidases [Bacteroides thetaiotaomicron VPI-5482],2VL4_A Structural and biochemical evidence for a boat-like transition state in beta-mannosidases [Bacteroides thetaiotaomicron VPI-5482],2VL4_B Structural and biochemical evidence for a boat-like transition state in beta-mannosidases [Bacteroides thetaiotaomicron VPI-5482],2VMF_A Structural and biochemical evidence for a boat-like transition state in beta-mannosidases [Bacteroides thetaiotaomicron VPI-5482],2VMF_B Structural and biochemical evidence for a boat-like transition state in beta-mannosidases [Bacteroides thetaiotaomicron VPI-5482],2VO5_A Structural and biochemical evidence for a boat-like transition state in beta-mannosidases [Bacteroides thetaiotaomicron VPI-5482],2VO5_B Structural and biochemical evidence for a boat-like transition state in beta-mannosidases [Bacteroides thetaiotaomicron VPI-5482],2VOT_A Structural and biochemical evidence for a boat-like transition state in beta-mannosidases [Bacteroides thetaiotaomicron VPI-5482],2VOT_B Structural and biochemical evidence for a boat-like transition state in beta-mannosidases [Bacteroides thetaiotaomicron VPI-5482],2VQT_A Structural and biochemical evidence for a boat-like transition state in beta-mannosidases [Bacteroides thetaiotaomicron],2VQT_B Structural and biochemical evidence for a boat-like transition state in beta-mannosidases [Bacteroides thetaiotaomicron],2VR4_A Transition-state mimicry in mannoside hydrolysis: characterisation of twenty six inhibitors and insight into binding from linear free energy relationships and 3-D structure [Bacteroides thetaiotaomicron VPI-5482],2VR4_B Transition-state mimicry in mannoside hydrolysis: characterisation of twenty six inhibitors and insight into binding from linear free energy relationships and 3-D structure [Bacteroides thetaiotaomicron VPI-5482] |
| 2JE8_A | 6.73e-194 | 30 | 831 | 13 | 816 | Structureof a beta-mannosidase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482],2JE8_B Structure of a beta-mannosidase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482] |
| 7OP6_A | 6.94e-194 | 30 | 831 | 13 | 816 | ChainA, Beta-mannosidase [Bacteroides thetaiotaomicron VPI-5482],7OP6_B Chain B, Beta-mannosidase [Bacteroides thetaiotaomicron VPI-5482],7OP7_A Chain A, Beta-mannosidase [Bacteroides thetaiotaomicron VPI-5482],7OP7_B Chain B, Beta-mannosidase [Bacteroides thetaiotaomicron VPI-5482] |
| 2WBK_A | 1.83e-193 | 30 | 831 | 11 | 814 | Structureof the Michaelis complex of beta-mannosidase, Man2A, provides insight into the conformational itinerary of mannoside hydrolysis [Bacteroides thetaiotaomicron VPI-5482],2WBK_B Structure of the Michaelis complex of beta-mannosidase, Man2A, provides insight into the conformational itinerary of mannoside hydrolysis [Bacteroides thetaiotaomicron VPI-5482] |
| 2VQU_A | 7.88e-192 | 30 | 831 | 11 | 814 | Structuraland biochemical evidence for a boat-like transition state in beta-mannosidases [Bacteroides thetaiotaomicron],2VQU_B Structural and biochemical evidence for a boat-like transition state in beta-mannosidases [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q95327 | 6.92e-124 | 28 | 676 | 23 | 692 | Beta-mannosidase OS=Capra hircus OX=9925 GN=MANBA PE=1 SV=1 |
| Q29444 | 5.45e-122 | 9 | 676 | 5 | 692 | Beta-mannosidase OS=Bos taurus OX=9913 GN=MANBA PE=1 SV=1 |
| Q5B7W2 | 6.45e-114 | 30 | 834 | 9 | 827 | Beta-mannosidase B OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=mndB PE=1 SV=2 |
| A1CGA8 | 7.29e-112 | 27 | 683 | 6 | 659 | Beta-mannosidase B OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=mndB PE=3 SV=1 |
| A1D911 | 2.77e-111 | 30 | 709 | 9 | 703 | Beta-mannosidase B OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=mndB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
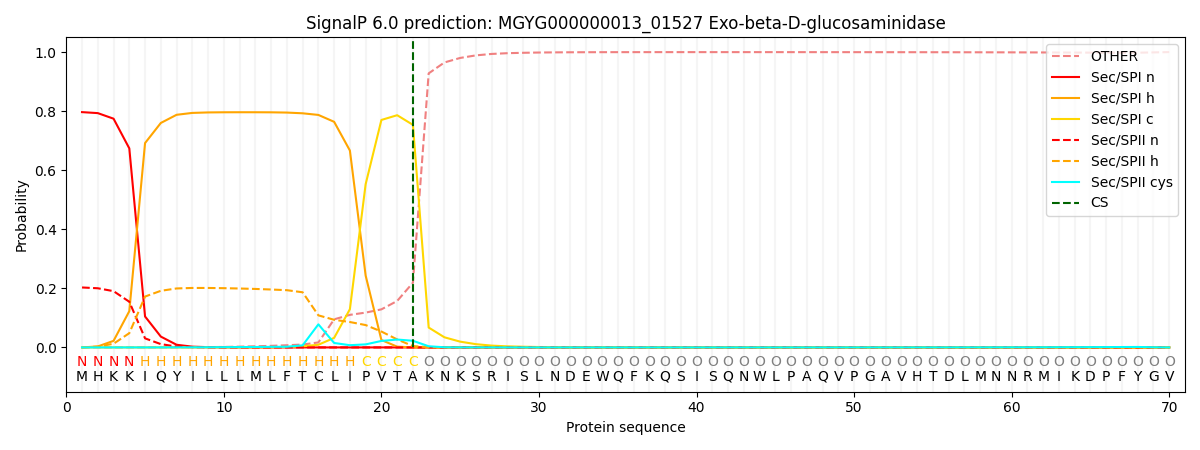
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001003 | 0.786475 | 0.211686 | 0.000301 | 0.000262 | 0.000243 |