You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000013_04537
You are here: Home > Sequence: MGYG000000013_04537
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides sp902362375 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides sp902362375 | |||||||||||
| CAZyme ID | MGYG000000013_04537 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 46712; End: 49984 Strand: + | |||||||||||
Full Sequence Download help
| MIIIESDRMK MKKQLIIIFL LCLAAKGISQ IPFDGSDSYA TPPIGFGQSE FENPLINSIN | 60 |
| REPYGATSIS FPTETEALQV KRSSSSRYQS LNGTWKFKFI TDWDNLPSDF MKVETNDDSW | 120 |
| DNIPVPSTWE MKGYGDQVYC GQGYEFRPVN PPFVPRKDNH IALYRKTFEV PASWEGQNVL | 180 |
| IHFAGVRGAF YLYVNGKKVG YNEDGGTLPA VFDMTPFLKR GKNQLAVQVL RWSDGSYLED | 240 |
| QDHWRFHGIT RDVYIESRPD VFIQDFAVIT DLDKDYKDAK LRIRPVISSK KTVDVSDWVL | 300 |
| EARLYTKEGN PALGVDMSMP VKTITTEKYQ QNFSLAKYLE TNVKAPLLWS SETPNLYIIV | 360 |
| LTLKDNKGNV VESRSSRIGF RKVEFKNNHE LCVNGKREYI YGVNRHDHDA WEGKTVPYER | 420 |
| MVQDVTLMKQ FGFNSVRTSH YPADPAFYDL CDEYGIYVMD EANVETCGAD AELSNNELWL | 480 |
| FAQMERVAGM VKRDKNHPSI IFWSLGNESG VGANNAARAS WVKDYDPTRL VHFEAYMHNG | 540 |
| GSRQYGYGID FMKTNRPAVN PPEPPAVDVV STMYPSVEGI IKLATQEGET RPVLMCEYAH | 600 |
| AKGNALGNHQ EYWNAVKKYP RLIGGYIWDW VDQSVIRKDD VTGKEYFSSL NGTNGLVFAD | 660 |
| RKIKPAINEC KKIYQHIRFD YTNGELTIKN EYNYLPLSAF NFSWKLMLNG ELIKNGELTD | 720 |
| IEAFPGTSAR VKIDTGYPDS GQKGEVILEI SAYLKKGTIW APKGFEIAWE QFTLQEGKVN | 780 |
| PTVEGNTGNG GKLTVQKKAN EIGIYNDRIN IVFNKKTGLI QSWIVGSKEF LERGPRINLW | 840 |
| RAPTHNDGGY RPKTENEISR QWVEAGLDSL QHKLKSFKLT EEKNGTVSVA TTFIAQKTGN | 900 |
| KSYVEYITKY IVDSSGKVQI DTDLKPFGNI ISFPRIGYTM TVKSGNDTFS WYGYGPYDTY | 960 |
| NDRHSGARLG KFSGTVDEQF THHAYPQENG NKYHCSWVSL TDNEGIGLIV EGMPFIESSV | 1020 |
| MHYSLENLSE AIDESQLKRT DNVTWNIDYK TYPIGNRSCG PPPLEQYVLF AEPVSFSFCI | 1080 |
| YPVLNQKAVQ | 1090 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 76 | 969 | 5.9e-222 | 0.976063829787234 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK09525 | lacZ | 0.0 | 52 | 1048 | 18 | 991 | beta-galactosidase. |
| PRK10340 | ebgA | 0.0 | 52 | 1086 | 5 | 1003 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| COG3250 | LacZ | 1.15e-151 | 85 | 971 | 9 | 808 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| pfam02836 | Glyco_hydro_2_C | 4.05e-89 | 393 | 679 | 10 | 302 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| smart01038 | Bgal_small_N | 7.98e-85 | 811 | 1080 | 7 | 272 | Beta galactosidase small chain. This domain comprises the small chain of dimeric beta-galactosidases EC:3.2.1.23. This domain is also found in single chain beta-galactosidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AWX43714.1 | 1.15e-264 | 42 | 1083 | 18 | 1056 |
| QGY43181.1 | 8.33e-253 | 49 | 1090 | 21 | 1102 |
| AWB67412.1 | 4.25e-251 | 42 | 1080 | 26 | 1077 |
| ARV16609.1 | 8.99e-249 | 48 | 1082 | 23 | 1074 |
| QVY66603.1 | 2.65e-247 | 48 | 1090 | 21 | 1078 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6S6Z_A | 4.70e-219 | 50 | 1078 | 3 | 977 | Structureof beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_B Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_C Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_D Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_E Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_F Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_G Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_H Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8] |
| 6SD0_A | 4.84e-219 | 50 | 1078 | 4 | 978 | Structureof beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_B Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_C Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_D Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8] |
| 3BGA_A | 1.94e-168 | 85 | 1083 | 45 | 1001 | Crystalstructure of beta-galactosidase from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482],3BGA_B Crystal structure of beta-galactosidase from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482] |
| 3DEC_A | 2.38e-166 | 85 | 1083 | 41 | 997 | ChainA, Beta-galactosidase [Bacteroides thetaiotaomicron VPI-5482] |
| 1JZ7_A | 1.28e-164 | 43 | 1065 | 8 | 1004 | E.COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH GALACTOSE [Escherichia coli],1JZ7_B E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH GALACTOSE [Escherichia coli],1JZ7_C E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH GALACTOSE [Escherichia coli],1JZ7_D E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH GALACTOSE [Escherichia coli],4TTG_A Beta-galactosidase (E. coli) in the presence of potassium chloride. [Escherichia coli],4TTG_B Beta-galactosidase (E. coli) in the presence of potassium chloride. [Escherichia coli],4TTG_C Beta-galactosidase (E. coli) in the presence of potassium chloride. [Escherichia coli],4TTG_D Beta-galactosidase (E. coli) in the presence of potassium chloride. [Escherichia coli] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q56307 | 2.65e-218 | 50 | 1078 | 4 | 978 | Beta-galactosidase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=lacZ PE=1 SV=2 |
| O52847 | 5.71e-209 | 39 | 1055 | 10 | 1004 | Beta-galactosidase OS=Priestia megaterium (strain DSM 319 / IMG 1521) OX=592022 GN=bgaM PE=3 SV=1 |
| Q6LL68 | 2.50e-177 | 89 | 1085 | 47 | 1030 | Beta-galactosidase OS=Photobacterium profundum (strain SS9) OX=298386 GN=lacZ PE=3 SV=1 |
| A1SWB8 | 1.11e-176 | 84 | 1072 | 44 | 1024 | Beta-galactosidase OS=Psychromonas ingrahamii (strain 37) OX=357804 GN=lacZ PE=3 SV=1 |
| P81650 | 7.26e-175 | 50 | 1080 | 12 | 1033 | Beta-galactosidase OS=Pseudoalteromonas haloplanktis OX=228 GN=lacZ PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
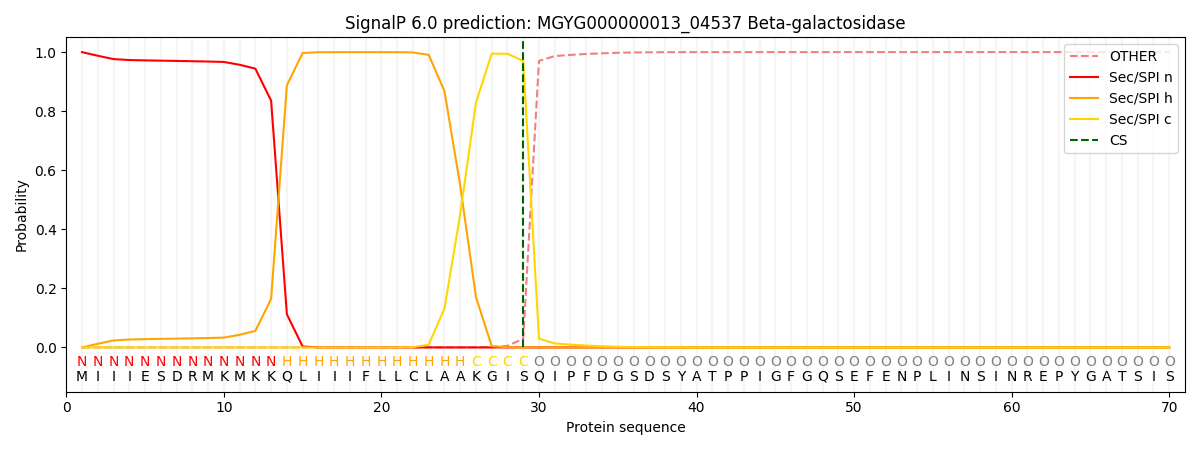
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000280 | 0.999062 | 0.000212 | 0.000144 | 0.000142 | 0.000129 |
