You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000054_00009
You are here: Home > Sequence: MGYG000000054_00009
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides acidifaciens | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides acidifaciens | |||||||||||
| CAZyme ID | MGYG000000054_00009 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Evolved beta-galactosidase subunit alpha | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 13844; End: 17185 Strand: + | |||||||||||
Full Sequence Download help
| MGIFSLVTTN TIADNAQKPY WQDMQVVAVN KEYPRSSFMT YDNRNDAMSG KFERSKYYRL | 60 |
| LNGTWKFYFA DSYKDLPDNI TDPNTSTDSW YDIQVPGNWE VQGHGVAIYT NHGYEFKARN | 120 |
| PQPPILPEAN PVGVYRRDID IPADWDGRDI YLHLAGAKSG VYVYINGKEV GYSEDSKNPA | 180 |
| EFLINPYVKP GKNVLTLKIF RWSTGSYLEC QDFWRISGIE RDVFVYSQPK AALKDFRVTS | 240 |
| TLDDSYKNGI FDLNVDLRNH KKEAANLTLV YELLDAQGKV IATEEKTAYI PTDEVRTLSF | 300 |
| DKKLADVNTW TSEHPNLYKL LMTVKENGKV KEIIPFNVGF RRIEIKPIEQ KAANGKPYVC | 360 |
| LFINGQPLKL KGVNIHEHNP ATGHYVTEEL MRRDFELMKQ HNLNSVRLCH YPQDRRFYEL | 420 |
| CDEYGLYVYD EANIESHGMY YDLRKGGSLG NNPEWLKLHM DRTINMFERN KNYPSVTFWS | 480 |
| LGNEAGNGYN FYQTYLWLKE ADKNIMARPV NYERAQWEWN TDMYVPQYPG ADWLENIGKN | 540 |
| GSDRPIAPSE YAHAMGNSTG NLWGQWQAIY KYPNLQGGYI WDWVDQGILQ KDKNGREYWA | 600 |
| YGGDFGVNMP SDGNFLCNGL VNPDRGPHPA MAEVKYVHQN VGFEATDAAS GKFKITNRFY | 660 |
| FTNLKKYQIH YNVVANGKNI RGGKVSLDIA PQASKEFAVP INGLKAQPGT EYFVNFSVTT | 720 |
| TEPEPLIPVG YEIAYDQFQL PIHAEKGTYK VSGPALNTST QGDELIASSS KVNFVFNKKS | 780 |
| GLVTSYKVDG TEYFKDGFGI QPNFWRAPND NDYGNGAPKR LQVWKQSSKN FHVTDASMTT | 840 |
| ENKVVSLKVT YLLAAGNLYV VTYKIYPSGA VNVNATFTST DMQAAETEVS EATRMATFTP | 900 |
| GSGAARKAAS KLEVPRIGVR FRLPAKMNNV QYFGRGPEEN YIDRNHGTLV GVYKTTADKM | 960 |
| YFNYVRPQEN GHHTDTRWIA LSPDKGSGLA IVADSLVGFN ALRNSIEDFD SEEALPHPYQ | 1020 |
| WNNFSPEEVA NHDEQAARNV LRRMHHVNDI TPRDFVEVCV DMKQQGVGGY DSWGAHPEPF | 1080 |
| HQIPANREYN WGFTLVPVRS AGQANEAAKY EYQ | 1113 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 46 | 949 | 9.1e-211 | 0.9747340425531915 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK09525 | lacZ | 0.0 | 21 | 1095 | 17 | 1024 | beta-galactosidase. |
| PRK10340 | ebgA | 0.0 | 21 | 1112 | 4 | 1015 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| COG3250 | LacZ | 8.88e-147 | 61 | 944 | 15 | 800 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| smart01038 | Bgal_small_N | 1.40e-102 | 768 | 1095 | 1 | 272 | Beta galactosidase small chain. This domain comprises the small chain of dimeric beta-galactosidases EC:3.2.1.23. This domain is also found in single chain beta-galactosidase. |
| pfam02836 | Glyco_hydro_2_C | 6.52e-90 | 361 | 643 | 8 | 302 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBK68335.1 | 0.0 | 1 | 1113 | 1 | 1113 |
| QUT33020.1 | 0.0 | 1 | 1113 | 1 | 1113 |
| QUR46524.1 | 0.0 | 12 | 1113 | 1 | 1102 |
| SCV08523.1 | 0.0 | 1 | 1113 | 15 | 1127 |
| QRQ55912.1 | 0.0 | 1 | 1113 | 15 | 1127 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6S6Z_A | 3.81e-235 | 19 | 1095 | 1 | 979 | Structureof beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_B Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_C Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_D Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_E Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_F Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_G Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_H Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8] |
| 6SD0_A | 3.93e-235 | 19 | 1095 | 2 | 980 | Structureof beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_B Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_C Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_D Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8] |
| 3DEC_A | 1.12e-219 | 17 | 1099 | 3 | 998 | ChainA, Beta-galactosidase [Bacteroides thetaiotaomicron VPI-5482] |
| 3BGA_A | 3.82e-213 | 19 | 1099 | 8 | 1002 | Crystalstructure of beta-galactosidase from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482],3BGA_B Crystal structure of beta-galactosidase from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482] |
| 1F4A_A | 6.09e-142 | 9 | 988 | 2 | 951 | E.COLI (LACZ) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-ORTHORHOMBIC) [Escherichia coli],1F4A_B E. COLI (LACZ) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-ORTHORHOMBIC) [Escherichia coli],1F4A_C E. COLI (LACZ) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-ORTHORHOMBIC) [Escherichia coli],1F4A_D E. COLI (LACZ) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-ORTHORHOMBIC) [Escherichia coli],1F4H_A E. COLI (LACZ) BETA-GALACTOSIDASE (ORTHORHOMBIC) [Escherichia coli],1F4H_B E. COLI (LACZ) BETA-GALACTOSIDASE (ORTHORHOMBIC) [Escherichia coli],1F4H_C E. COLI (LACZ) BETA-GALACTOSIDASE (ORTHORHOMBIC) [Escherichia coli],1F4H_D E. COLI (LACZ) BETA-GALACTOSIDASE (ORTHORHOMBIC) [Escherichia coli] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q56307 | 2.15e-234 | 19 | 1095 | 2 | 980 | Beta-galactosidase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=lacZ PE=1 SV=2 |
| O52847 | 1.95e-213 | 19 | 1098 | 18 | 1034 | Beta-galactosidase OS=Priestia megaterium (strain DSM 319 / IMG 1521) OX=592022 GN=bgaM PE=3 SV=1 |
| A5F5U6 | 1.35e-173 | 21 | 1103 | 13 | 1024 | Beta-galactosidase OS=Vibrio cholerae serotype O1 (strain ATCC 39541 / Classical Ogawa 395 / O395) OX=345073 GN=lacZ PE=3 SV=2 |
| Q9K9C6 | 8.32e-167 | 22 | 1095 | 11 | 1011 | Beta-galactosidase OS=Alkalihalobacillus halodurans (strain ATCC BAA-125 / DSM 18197 / FERM 7344 / JCM 9153 / C-125) OX=272558 GN=lacZ PE=3 SV=1 |
| Q6LL68 | 4.90e-166 | 12 | 1097 | 3 | 1027 | Beta-galactosidase OS=Photobacterium profundum (strain SS9) OX=298386 GN=lacZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
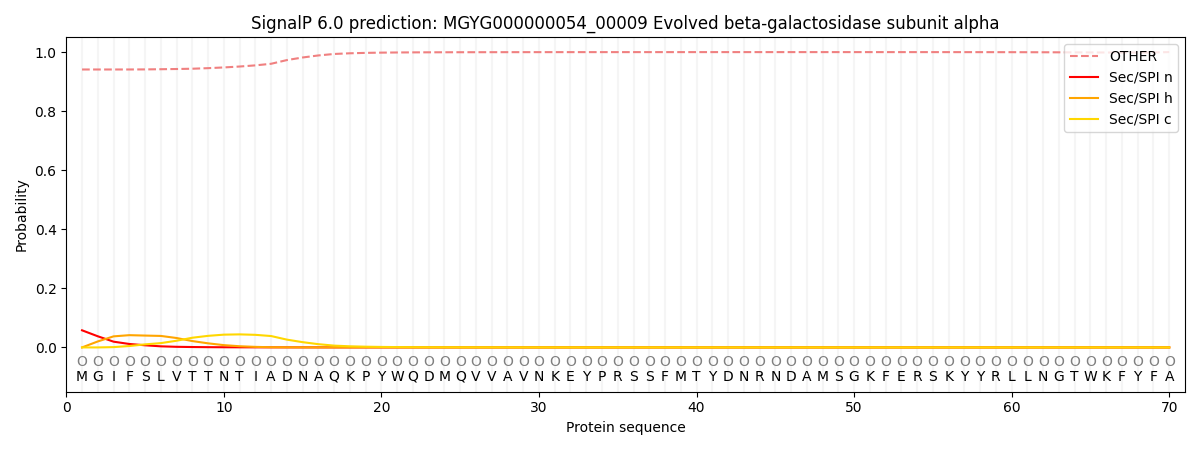
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.945491 | 0.052967 | 0.000831 | 0.000202 | 0.000105 | 0.000406 |
