You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000284_00025
You are here: Home > Sequence: MGYG000000284_00025
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parabacteroides sp900540715 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides sp900540715 | |||||||||||
| CAZyme ID | MGYG000000284_00025 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase BoGH2A | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 37326; End: 39983 Strand: - | |||||||||||
Full Sequence Download help
| MKKNILVLLF WVLTFNLSAS GSPEFSTAGF YELPNTGREV YSMNLAWRFI KGDVSGTPYA | 60 |
| ADFDDSGWEV VSLPHGLEYL PVEASGCANY QGVAWYRKRF TPDDRLKGKQ LFVHFEAIMG | 120 |
| KSEVYVNGQL LKTHYGGYLP VILDVTNLLK WDEENVIAVK ADNSDDPSYP VGKPQNLLDF | 180 |
| TYFGGIYRDC WLVAHNDVFI TNSNYEDEVA GGGLFVSYAD VSEKSALVNL KAHIRNHSAS | 240 |
| DKKVTVTFEL KKKSGEIVRR VSTGLAVKRG DASYAVGSMK INTPSLWSPD HPYLYDLEVM | 300 |
| VRGANGQVLD GYRQRVGIRS IEFKQTEGLW INGKPYEGKL MGTNRHQDFA VLGNALPNSM | 360 |
| QWRDAKKLRD AGMKVIRTHY VIDPAFMDAC DELGLFALVE VPGWQFWNKE PVFSERVYSD | 420 |
| IRNMIRIHRN HASLFFWEPI LNETHYPEEF AKKALDICQE EYPYPYSIAA CDNGAAGDHY | 480 |
| YSLLLRPIES KLRTDKTYFI REWGDNVDDW NAQNSDSRVA RGWGEVPMLL QAEHYANPSY | 540 |
| VKEYPVLCYE TICNKPSQIV GACFWHSFDH QRGYHADPFY GGIMDAFRQP KSSYYMFMAQ | 600 |
| RSPEKSDLIA DNGPMIYIAH EITPFSPEDV TVYSNCDEVR LTVFKDGKEY KYKKDPNHKG | 660 |
| MPSPIITFKD VFHFMEWKTM ARSGKQNDAY LLAEGLIDGK VVVSHKRYPS GQADHIALRL | 720 |
| DHEQVSLKAD GSDVVTVIAE VVDKRGTVKR LNNSCIRFQI EGEGHILGDA SVGTNPCPLI | 780 |
| WGTAPVLVQS TANPGKIKII ASMQNPGESR PLEGVLEFES VVNDQKEIYS EKELASSGQP | 840 |
| FVVTDRSVNK SDLERENERL RKELNQLKVK EVEKQQTQFG VGIND | 885 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 35 | 604 | 5e-91 | 0.6183510638297872 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 4.75e-50 | 92 | 450 | 82 | 413 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 9.34e-42 | 38 | 443 | 10 | 417 | beta-D-glucuronidase; Provisional |
| PRK10340 | ebgA | 2.07e-34 | 96 | 456 | 113 | 467 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| pfam00703 | Glyco_hydro_2 | 1.82e-17 | 214 | 319 | 6 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| PRK09525 | lacZ | 4.22e-16 | 79 | 437 | 114 | 457 | beta-galactosidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ALJ59608.1 | 0.0 | 5 | 880 | 10 | 890 |
| QUT89356.1 | 0.0 | 5 | 880 | 8 | 888 |
| QDO67981.1 | 0.0 | 5 | 880 | 8 | 888 |
| QUT75446.1 | 0.0 | 1 | 880 | 1 | 885 |
| QJR74048.1 | 0.0 | 1 | 880 | 1 | 889 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5UJ6_A | 3.62e-61 | 20 | 437 | 7 | 413 | CrystalStructure of Bacteroides Uniformis beta-glucuronidase [Bacteroides uniformis str. 3978 T3 ii],5UJ6_B Crystal Structure of Bacteroides Uniformis beta-glucuronidase [Bacteroides uniformis str. 3978 T3 ii],6NZG_A Bacteroides uniformis beta-glucuronidase 2 covalently bound to cyclophellitol-6-carboxylate aziridine [Bacteroides uniformis],6NZG_B Bacteroides uniformis beta-glucuronidase 2 covalently bound to cyclophellitol-6-carboxylate aziridine [Bacteroides uniformis] |
| 6D50_A | 1.50e-59 | 38 | 437 | 33 | 421 | Bacteroidesuniforms beta-glucuronidase 2 bound to D-glucaro-1,5-lactone [Bacteroides uniformis str. 3978 T3 ii],6D50_B Bacteroides uniforms beta-glucuronidase 2 bound to D-glucaro-1,5-lactone [Bacteroides uniformis str. 3978 T3 ii] |
| 3FN9_A | 7.16e-59 | 38 | 649 | 5 | 619 | Crystalstructure of putative beta-galactosidase from bacteroides fragilis [Bacteroides fragilis NCTC 9343],3FN9_B Crystal structure of putative beta-galactosidase from bacteroides fragilis [Bacteroides fragilis NCTC 9343],3FN9_C Crystal structure of putative beta-galactosidase from bacteroides fragilis [Bacteroides fragilis NCTC 9343],3FN9_D Crystal structure of putative beta-galactosidase from bacteroides fragilis [Bacteroides fragilis NCTC 9343] |
| 6D8G_A | 4.13e-58 | 38 | 437 | 33 | 421 | D341AD367A calcium binding mutant of Bacteroides uniformis beta-glucuronidase 2 [Bacteroides uniformis str. 3978 T3 ii],6D8G_B D341A D367A calcium binding mutant of Bacteroides uniformis beta-glucuronidase 2 [Bacteroides uniformis str. 3978 T3 ii] |
| 6HPD_A | 8.91e-58 | 33 | 807 | 30 | 794 | Thestructure of a beta-glucuronidase from glycoside hydrolase family 2 [Formosa agariphila KMM 3901] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P77989 | 1.17e-105 | 37 | 797 | 2 | 719 | Beta-galactosidase OS=Thermoanaerobacter pseudethanolicus (strain ATCC 33223 / 39E) OX=340099 GN=lacZ PE=3 SV=2 |
| A7LXS9 | 2.97e-57 | 38 | 804 | 42 | 837 | Beta-galactosidase BoGH2A OS=Bacteroides ovatus (strain ATCC 8483 / DSM 1896 / JCM 5824 / BCRC 10623 / CCUG 4943 / NCTC 11153) OX=411476 GN=BACOVA_02645 PE=1 SV=1 |
| T2KN75 | 4.62e-57 | 33 | 807 | 21 | 785 | Beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22060 PE=1 SV=1 |
| P26257 | 1.67e-56 | 38 | 772 | 2 | 710 | Beta-galactosidase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=lacZ PE=1 SV=1 |
| T2KPJ7 | 6.49e-48 | 26 | 746 | 39 | 754 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_21970 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
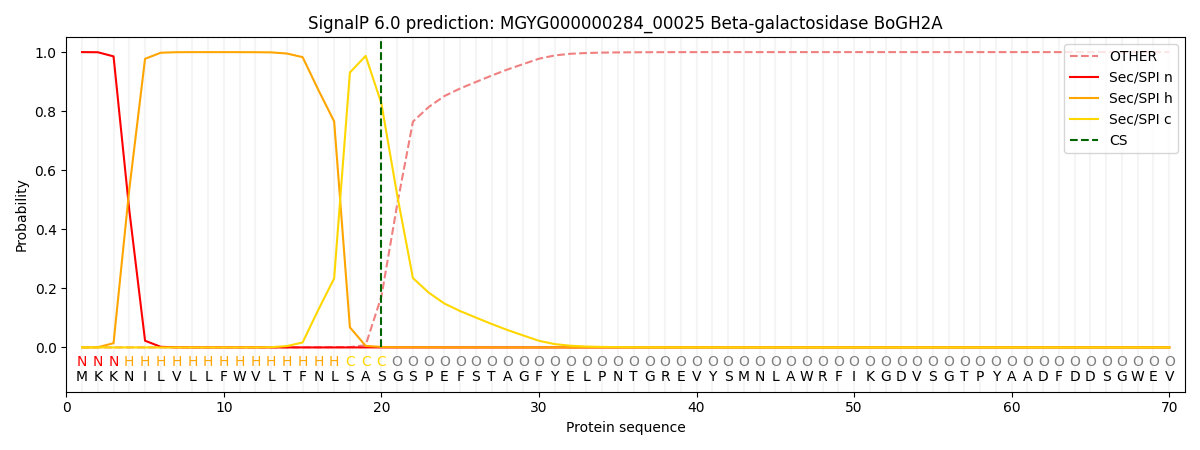
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000367 | 0.998881 | 0.000278 | 0.000167 | 0.000147 | 0.000145 |
