You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000534_00077
You are here: Home > Sequence: MGYG000000534_00077
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900543975 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900543975 | |||||||||||
| CAZyme ID | MGYG000000534_00077 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 99090; End: 102701 Strand: - | |||||||||||
Full Sequence Download help
| MNTITNLKLT AVALTALTLT SFTDGKTRGI GVYPGNPAES YAPSLRTDNA YRNVALHRIV | 60 |
| RTSSQHDLNL TPQLLTDGII ATNSPQYLVA TTNSGVLQNR ERESGIDGNE YTNMNLMGTK | 120 |
| AWLQYRWNNM LIKADKVKII CTVAYKPAEA KKGYSVKVSV DNKVVAEETG KELPGKESKQ | 180 |
| RVSSDPNKQT ASGELPLRIV EMELPIGKSG KVEFDNLRFN FDMAGAAYWT VTEIKFFNNG | 240 |
| QAVTDLQPAR DFTSAWMSDK GGQQWAYVDL GTAAEIDNVK LSWIEKAVKG RVDVATEMGH | 300 |
| WTTVAQLPGG KALADNLKFE PVKARFVRVL MTQPTATGRY IMSEMEVMGR GGLVAQPHAL | 360 |
| GGMKAGRYLL NGGSWQVQRA SEVKGSGEQI SQAAFNTNGW VAATVPGTVL TSYVNVGALP | 420 |
| DPALADNWNN ASESFFNSNF WYRTAFNLPT EMKGKHVFLN FDGINWKANV YLNGKKIDRI | 480 |
| EGAFTRSKVD ITNLLNANGA NILAVEIVKN AHPGSIKVKN RKNTDFNGGI LGYDNPTFHA | 540 |
| TIGWDWITTV RGRDIGIWND VALTTAGDVS LSDPVVTTTL TGADNKASMT PTVVLANNEA | 600 |
| REVKGELHGW IGNISFSKTV TLAPNATQEV SFSPDEFAQL KEQNMKLWWP NGYGTPYLYK | 660 |
| AGFEFATDEG VQSRTCYKAG IRQMTYKDID TRLTMYINGK RFIPLGGNWG FSEQNLLYRG | 720 |
| REYDIAVRYH RDMNFNMIRN WVGMTGDEEF YEACDKYGIM VWQDFWLANP CDGPDPQNED | 780 |
| MFLANARDYT LRMRQHPSIG LYCGRNEGYP PATIDKALRQ YVAAMNPGMG YISSSADDGV | 840 |
| SGHGPYWACT PKFYFEHQTG KLHSERGMPN VMTFEGLSRT LAPNALWPQG DAWGEHDYGM | 900 |
| EGAQRGASFN GIIEKAFGKV TDARQFTALA QWENYDGYRA MYESGSKDRM GLIIWMSHSC | 960 |
| WPSMTWCCYD YYFEPTAAFF GSKKACEPLH IQLNASTRNP EVVNLAAGEH KQLTAKREVI | 1020 |
| ALDGKTVKED VATLDTNDDT TTPLQALHVD DSYDLGGKNV YIVRLTLSDS NGKVLSTNTY | 1080 |
| VDSNDHGNLQ DLNALPVLGE NDIKVSVETV ACKDAKKACC SAQAAGCNKS RRIITLENTS | 1140 |
| KTPAMMVRLN LLDGDGDQIL PVDYSDNYFH LLPGEKRTIK VAWNNEDMRK GNPVVDVTGF | 1200 |
| NVK | 1203 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 368 | 1044 | 1.8e-85 | 0.6861702127659575 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 1.40e-35 | 423 | 819 | 46 | 418 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| pfam18368 | Ig_GlcNase | 1.11e-30 | 1078 | 1202 | 1 | 104 | Exo-beta-D-glucosaminidase Ig-fold domain. This domain can be found in 2 glycoside hydrolase subfamily of beta-glucosaminidases (EC:3.2.1.165) such as CsxA, from Amycolatopsis orientalis that has exo-beta-D-glucosaminidase (exo-chitosanase) activity. It has an immunoglobulin-like topology. |
| PRK10150 | PRK10150 | 1.69e-13 | 439 | 821 | 66 | 428 | beta-D-glucuronidase; Provisional |
| PRK10340 | ebgA | 2.70e-10 | 442 | 807 | 113 | 449 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| pfam00703 | Glyco_hydro_2 | 3.85e-09 | 569 | 682 | 1 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ALO48125.1 | 0.0 | 27 | 1202 | 27 | 1175 |
| QUR48317.1 | 0.0 | 24 | 1203 | 43 | 1207 |
| BBK93008.1 | 0.0 | 24 | 1203 | 43 | 1207 |
| AST55694.1 | 0.0 | 24 | 1202 | 43 | 1206 |
| QRO15146.1 | 0.0 | 24 | 1202 | 43 | 1206 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2VZO_A | 8.52e-62 | 351 | 1201 | 35 | 890 | Crystalstructure of Amycolatopsis orientalis exo-chitosanase CsxA [Amycolatopsis orientalis],2VZO_B Crystal structure of Amycolatopsis orientalis exo-chitosanase CsxA [Amycolatopsis orientalis] |
| 2VZT_A | 2.02e-61 | 351 | 1201 | 35 | 890 | Complexof Amycolatopsis orientalis exo-chitosanase CsxA E541A with PNP-beta-D-glucosamine [Amycolatopsis orientalis],2VZT_B Complex of Amycolatopsis orientalis exo-chitosanase CsxA E541A with PNP-beta-D-glucosamine [Amycolatopsis orientalis],2VZV_A Substrate Complex of Amycolatopsis orientalis exo-chitosanase CsxA E541A with chitosan [Amycolatopsis orientalis],2VZV_B Substrate Complex of Amycolatopsis orientalis exo-chitosanase CsxA E541A with chitosan [Amycolatopsis orientalis] |
| 2VZS_A | 2.02e-61 | 351 | 1201 | 35 | 890 | ChitosanProduct complex of Amycolatopsis orientalis exo-chitosanase CsxA [Amycolatopsis orientalis],2VZS_B Chitosan Product complex of Amycolatopsis orientalis exo-chitosanase CsxA [Amycolatopsis orientalis],2X05_A Inhibition of the exo-beta-D-glucosaminidase CsxA by a glucosamine- configured castanospermine and an amino-australine analogue [Amycolatopsis orientalis],2X05_B Inhibition of the exo-beta-D-glucosaminidase CsxA by a glucosamine- configured castanospermine and an amino-australine analogue [Amycolatopsis orientalis],2X09_A Inhibition of the exo-beta-D-glucosaminidase CsxA by a glucosamine- configured castanospermine and an amino-australine analogue [Amycolatopsis orientalis],2X09_B Inhibition of the exo-beta-D-glucosaminidase CsxA by a glucosamine- configured castanospermine and an amino-australine analogue [Amycolatopsis orientalis] |
| 2VZU_A | 4.75e-60 | 351 | 1201 | 35 | 890 | Complexof Amycolatopsis orientalis exo-chitosanase CsxA D469A with PNP-beta-D-glucosamine [Amycolatopsis orientalis],2VZU_B Complex of Amycolatopsis orientalis exo-chitosanase CsxA D469A with PNP-beta-D-glucosamine [Amycolatopsis orientalis] |
| 6BYE_A | 7.07e-34 | 373 | 1182 | 10 | 833 | Crystalstructure of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri in complex with mannose [Xanthomonas citri pv. citri str. 306],6BYE_B Crystal structure of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri in complex with mannose [Xanthomonas citri pv. citri str. 306] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q5H7P5 | 1.47e-80 | 366 | 1201 | 2 | 944 | Mannosylglycoprotein endo-beta-mannosidase OS=Lilium longiflorum OX=4690 GN=EBM PE=1 SV=4 |
| Q75W54 | 3.22e-80 | 366 | 1201 | 5 | 935 | Mannosylglycoprotein endo-beta-mannosidase OS=Arabidopsis thaliana OX=3702 GN=EBM PE=1 SV=3 |
| Q82NR8 | 4.15e-74 | 375 | 1201 | 57 | 895 | Exo-beta-D-glucosaminidase OS=Streptomyces avermitilis (strain ATCC 31267 / DSM 46492 / JCM 5070 / NBRC 14893 / NCIMB 12804 / NRRL 8165 / MA-4680) OX=227882 GN=csxA PE=1 SV=1 |
| Q56F26 | 8.30e-61 | 351 | 1201 | 35 | 890 | Exo-beta-D-glucosaminidase OS=Amycolatopsis orientalis OX=31958 GN=csxA PE=1 SV=2 |
| Q4R1C4 | 3.71e-55 | 375 | 1203 | 38 | 886 | Exo-beta-D-glucosaminidase OS=Hypocrea jecorina OX=51453 GN=gls93 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
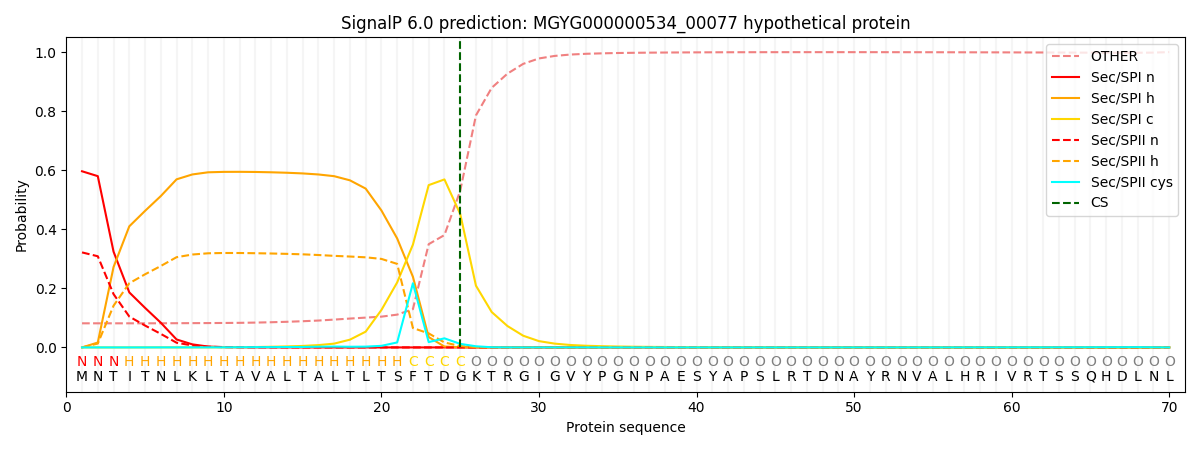
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.089430 | 0.583119 | 0.326048 | 0.000560 | 0.000399 | 0.000432 |
