You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001489_00114
You are here: Home > Sequence: MGYG000001489_00114
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parabacteroides goldsteinii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides goldsteinii | |||||||||||
| CAZyme ID | MGYG000001489_00114 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Evolved beta-galactosidase subunit alpha | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 128936; End: 132316 Strand: + | |||||||||||
Full Sequence Download help
| MKHLVKKAML TGAFGCFLLG TALADNATAS QDTKPYWQDV QVVAVNKELP RSSFMTYGDR | 60 |
| STALTSRFEK SPYYSLLNGT WKFYFVDSYK DLPANITDPS TSTSSWDDIT VPGNWELQGH | 120 |
| GTAIYTNHGY EFKPRNPQPP LLPETTPVGV YRRDFEIPAN WDGRDVYLHI AGAKSGLYVY | 180 |
| VNGKEVGYSE DSKNPAEFLI NDYLQPGKNV LTLKIFRWST GSYLECQDFW RISGIERDVF | 240 |
| LWSQPKIAVN DFRVISTLDD TYKNGIFNLA VDIKNHTKAA KDITVSYELL DAKGQTVATA | 300 |
| DNKLWVGANS IKTASFEKEL NDVATWSAEH PNLYKLLMTV KEDGKVTEVI PSNVGFRRIE | 360 |
| IKPIDQIAGN GKPYTVLLFN GQPIKFKGVN IHEHNPQTGH YMTEELMRKD FELMKKNNIN | 420 |
| SVRLCHYPQD RRFYELCDEY GLYVYDEANV ESHGMYYSLS KGGSLGNNPE WLLPHMDRTM | 480 |
| NMYERNKNYP SVTIWSLGNE AGNGYNFYQT YLYVKNKEKG AMNRPVNYER ALWEWNTDMY | 540 |
| VPQYPSAGWL EEIGKEGSDR PIAPSEYAHA MGNSTGNLWG QWKAIYKYPN LQGGWIWDWV | 600 |
| DQGLLVKDEN GKDYYAYGGD FGVNMPSDGN FLCNGIVNPD RTPHPAMKEV KFTHQNVGFD | 660 |
| AVDVANGVFK ITNRFYFTNL KDYTITYTVK ANNKVIKSDK VSLDLAPQAS QEITVPVAGL | 720 |
| KPQAGVDYLV NFVVTSNKQE GVIPAGYEIA QDQFRLPIEA DKVAYKAGGP KLAVSEEGDN | 780 |
| VKVTSSKVNF VFDKKSGVVS SYKVGGTEYF NEGFGIQPNF WRAPNDNDYG SGNPKRLEIW | 840 |
| ELSSRNFNVT KAAAEIVDGN AIVNVEYKLP AGNEFLITYK IYPDGVMNVA THFTPAHLDG | 900 |
| VKIGISEATA TATFSPGRDK VSDRDKMVVP RIGVRFRLPV TMDQLEYFGR GPVENYWDRK | 960 |
| AGYMIGQYKS TAEEQYFPYV RPQENGHHCD TRWLTLGGKG KNLLIVADDD MEFNAMRNSV | 1020 |
| EDFDAGKTTN RPYQWNNFTQ EEINNRPNIG PYDKPRQTHI NDITPRNFVE VCVDLKQQGL | 1080 |
| AGYDSWYSRP EPEYTLPADK EYKWGFTLIP AENASSAQKK AGYSYK | 1126 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 65 | 965 | 1.2e-205 | 0.9720744680851063 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK09525 | lacZ | 0.0 | 37 | 1108 | 17 | 1024 | beta-galactosidase. |
| PRK10340 | ebgA | 0.0 | 37 | 1126 | 4 | 1016 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| COG3250 | LacZ | 2.31e-143 | 60 | 960 | 3 | 801 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| smart01038 | Bgal_small_N | 1.05e-96 | 784 | 1108 | 1 | 272 | Beta galactosidase small chain. This domain comprises the small chain of dimeric beta-galactosidases EC:3.2.1.23. This domain is also found in single chain beta-galactosidase. |
| pfam02836 | Glyco_hydro_2_C | 4.71e-92 | 376 | 659 | 7 | 302 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT49251.1 | 0.0 | 1 | 1126 | 1 | 1125 |
| QUT53259.1 | 0.0 | 5 | 1110 | 4 | 1110 |
| QKH96585.1 | 0.0 | 5 | 1110 | 6 | 1112 |
| BBK89947.1 | 0.0 | 5 | 1110 | 6 | 1112 |
| QIX66118.1 | 0.0 | 5 | 1110 | 4 | 1110 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6S6Z_A | 4.72e-223 | 35 | 1116 | 1 | 987 | Structureof beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_B Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_C Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_D Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_E Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_F Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_G Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_H Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8] |
| 6SD0_A | 4.86e-223 | 35 | 1116 | 2 | 988 | Structureof beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_B Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_C Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_D Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8] |
| 3DEC_A | 1.23e-204 | 34 | 1112 | 4 | 998 | ChainA, Beta-galactosidase [Bacteroides thetaiotaomicron VPI-5482] |
| 3BGA_A | 7.74e-202 | 35 | 1112 | 8 | 1002 | Crystalstructure of beta-galactosidase from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482],3BGA_B Crystal structure of beta-galactosidase from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482] |
| 1JZ7_A | 1.42e-146 | 37 | 997 | 16 | 947 | E.COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH GALACTOSE [Escherichia coli],1JZ7_B E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH GALACTOSE [Escherichia coli],1JZ7_C E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH GALACTOSE [Escherichia coli],1JZ7_D E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH GALACTOSE [Escherichia coli],4TTG_A Beta-galactosidase (E. coli) in the presence of potassium chloride. [Escherichia coli],4TTG_B Beta-galactosidase (E. coli) in the presence of potassium chloride. [Escherichia coli],4TTG_C Beta-galactosidase (E. coli) in the presence of potassium chloride. [Escherichia coli],4TTG_D Beta-galactosidase (E. coli) in the presence of potassium chloride. [Escherichia coli] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O52847 | 6.02e-223 | 18 | 1110 | 1 | 1033 | Beta-galactosidase OS=Priestia megaterium (strain DSM 319 / IMG 1521) OX=592022 GN=bgaM PE=3 SV=1 |
| Q56307 | 2.66e-222 | 35 | 1116 | 2 | 988 | Beta-galactosidase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=lacZ PE=1 SV=2 |
| A5F5U6 | 4.57e-175 | 37 | 1108 | 13 | 1016 | Beta-galactosidase OS=Vibrio cholerae serotype O1 (strain ATCC 39541 / Classical Ogawa 395 / O395) OX=345073 GN=lacZ PE=3 SV=2 |
| Q9K9C6 | 3.48e-170 | 38 | 1108 | 11 | 1011 | Beta-galactosidase OS=Alkalihalobacillus halodurans (strain ATCC BAA-125 / DSM 18197 / FERM 7344 / JCM 9153 / C-125) OX=272558 GN=lacZ PE=3 SV=1 |
| A1SWB8 | 3.51e-168 | 37 | 1113 | 13 | 1035 | Beta-galactosidase OS=Psychromonas ingrahamii (strain 37) OX=357804 GN=lacZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
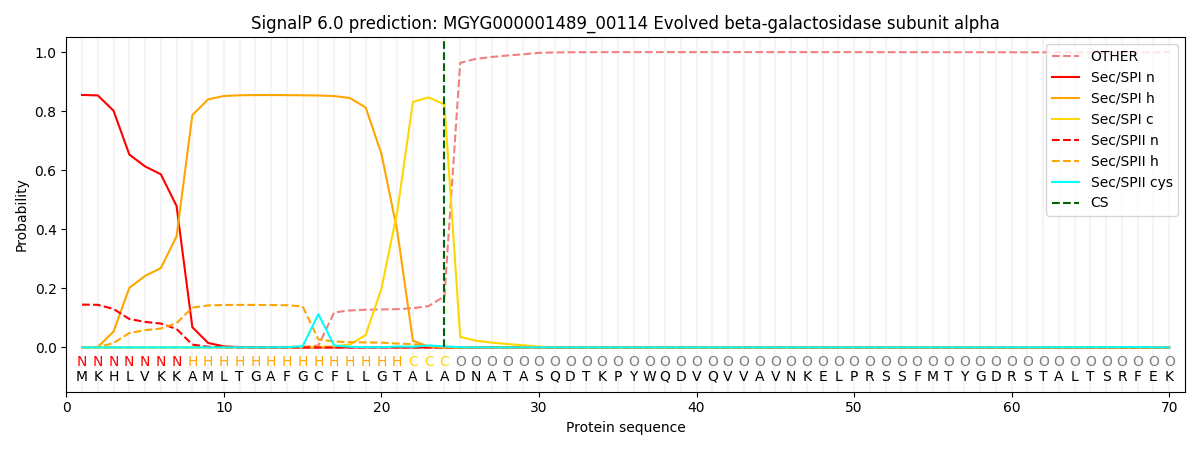
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000655 | 0.844823 | 0.153692 | 0.000295 | 0.000280 | 0.000233 |
