You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001771_00020
You are here: Home > Sequence: MGYG000001771_00020
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
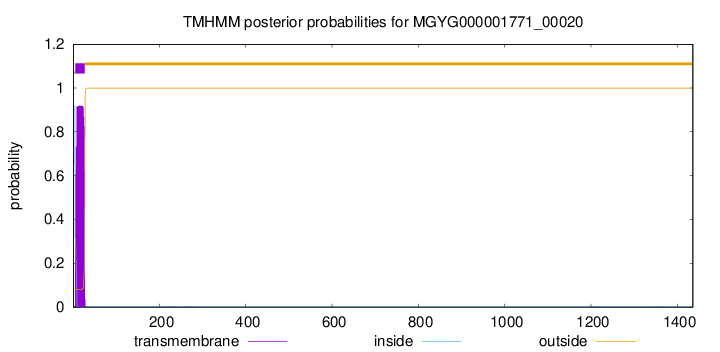
TMHMM annotations
Basic Information help
| Species | CAG-617 sp000438115 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; CAG-617; CAG-617 sp000438115 | |||||||||||
| CAZyme ID | MGYG000001771_00020 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 24623; End: 28933 Strand: - | |||||||||||
Full Sequence Download help
| MSKVFVMRAI WLLGVFLALM PSAVWAGDKV QAQHVYVITS VSQNKVLTNE DKKDNNVLIR | 60 |
| LAARDEQSLG QLWIVQAVDE SKSVYLLKNY QSGKALDMAD NGTGSSSFLQ WDVMPENANQ | 120 |
| QFCFVEHDNG TYLLQSAANP AKYAVLGSDN NSFTLGSDSQ AESFELTEVN VFVPMTDTRY | 180 |
| RIKSTSAEMY VSVKGSRADN SRIYTDAFNS DDKSQQWLLN ASANGSYSWL NMYSGKGIDL | 240 |
| AMEGVGYPLQ WSYQSSNANQ QFYFVNVPDL EGAVQMYGMQ GGVKHYLKAN GSQAMTETTD | 300 |
| STDPATYFIM EPMPNSERNH WEDQTFYEEN KERAHATFMP YASVEQMKAD EPYYSKPWNT | 360 |
| PAKAQYLSLN GIWNFKFVSS PEERPMEDFY GDDVDASKWD TISVPSCWEM KGYDKPLYVN | 420 |
| VEYPFADMPP YIFCKVDGIG ANPVGSYRRT FELPEGWDTQ RVFFHSDGLY SAAYVWVNGK | 480 |
| YVGYTQGGNN DAEFDITSYV RKGSNNISVQ VFRWSDASYL EGQDMFHMSG LHRDVYLYAT | 540 |
| PKTFVRDHYI TAQLDEASNY MSGNFNVDLE IDNRDAQAAK KAVEVTLYNP SGKKVKSWST | 600 |
| NVSLQDGDTI QHVSLGGQLD DLILWSAEKP NLYTVVVCQK NEKGKEEMAF STKYGFRQVE | 660 |
| IKNKLVYVNG QQVYFKGVNN QDTHPVYGRS IDVPTMLKDV TMMKQANMNT VRTSHYPRQA | 720 |
| KMYAMFDYYG LYVMDEADVE CHKNWDDHYY GTGCIARDPS WEGQFVDRNV RMVYRDRNHP | 780 |
| SVVFWSMGNE SQNGCNFTAA FKAMKAIDSR IIHYEGATRF PNGGDGADNT ELYSVMYPNL | 840 |
| TDVERNSNNN DRPYFMCEYA HAMGNAVGNL KEYWDIIESS KYGIGGCIWD WVDQSIIDAA | 900 |
| DIKNGQYTRN GFNYYRTGYD YPGPHQGNFV NNGLIQADRS WSAKLTEVKQ VYQYIKFKGF | 960 |
| SAASRQLTMT NKYDFTDLNE FELAYEVLKD GEVVESGKMD VPSTQPDANC VLTIPYKTEV | 1020 |
| AEGAEYLLNI NMLKKQATDW CDAGYSMASV QYMLQERPKT LPAKNDLTGK LKVTDNQSTV | 1080 |
| TISNEAGSFN VTFNNSNGTL TEYKVNNISY MNAAGSPQYD NYRWIENDTY GDNSNGIASV | 1140 |
| NTTRGLSTDG KTYTYKVVAV GSKCDYVLTY TIYLDGTIDM KADFTPKSAD LRRIGLSMNF | 1200 |
| LGGLDSVSYY ARGPWENYVD RKTGSMLGRY NTTTDKMFTP YPNPQTCGNR EDLREVSLTN | 1260 |
| PTTGNGLLIR TEGKVAFSLL PYTDQQMVNA RHPWELTRSS AVYAHFDYMQ RGLGNASCGP | 1320 |
| GTLSQYTCPS SGTYTYKLRF IPIINGEQVG VTPVVATDNA WTFRYDKGSE KVYCEGVANG | 1380 |
| RVQAMLVNQG GVRLSHREVN DNAPSQLAFS LNGQPNGSYL VVLKSANGQQ VYKFMK | 1436 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 359 | 1227 | 2e-194 | 0.9694148936170213 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK09525 | lacZ | 0.0 | 318 | 1340 | 14 | 1024 | beta-galactosidase. |
| PRK10340 | ebgA | 0.0 | 319 | 1347 | 2 | 1004 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| COG3250 | LacZ | 5.74e-141 | 368 | 1229 | 14 | 808 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| pfam02836 | Glyco_hydro_2_C | 2.56e-78 | 659 | 957 | 1 | 302 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| pfam02929 | Bgal_small_N | 1.55e-70 | 1085 | 1340 | 1 | 230 | Beta galactosidase small chain. This domain comprises the small chain of dimeric beta-galactosidases EC:3.2.1.23. This domain is also found in single chain beta-galactosidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QFQ12437.1 | 0.0 | 14 | 1349 | 13 | 1343 |
| ATS05896.1 | 0.0 | 305 | 1343 | 31 | 1068 |
| ATR96816.1 | 0.0 | 305 | 1343 | 31 | 1068 |
| ATS09286.1 | 0.0 | 305 | 1343 | 31 | 1068 |
| ATS03704.1 | 0.0 | 305 | 1343 | 31 | 1068 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6S6Z_A | 8.54e-189 | 321 | 1388 | 4 | 1022 | Structureof beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_B Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_C Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_D Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_E Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_F Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_G Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_H Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8] |
| 6SD0_A | 8.78e-189 | 321 | 1388 | 5 | 1023 | Structureof beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_B Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_C Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_D Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8] |
| 3BGA_A | 1.97e-169 | 309 | 1347 | 3 | 1005 | Crystalstructure of beta-galactosidase from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482],3BGA_B Crystal structure of beta-galactosidase from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482] |
| 3DEC_A | 1.55e-168 | 321 | 1346 | 7 | 1000 | ChainA, Beta-galactosidase [Bacteroides thetaiotaomicron VPI-5482] |
| 3MUY_1 | 3.75e-137 | 317 | 1338 | 12 | 1018 | Chain1, Beta-D-galactosidase [Escherichia coli K-12],3MUY_2 Chain 2, Beta-D-galactosidase [Escherichia coli K-12],3MUY_3 Chain 3, Beta-D-galactosidase [Escherichia coli K-12],3MUY_4 Chain 4, Beta-D-galactosidase [Escherichia coli K-12] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O52847 | 2.22e-189 | 330 | 1343 | 30 | 1034 | Beta-galactosidase OS=Priestia megaterium (strain DSM 319 / IMG 1521) OX=592022 GN=bgaM PE=3 SV=1 |
| Q56307 | 4.81e-188 | 321 | 1388 | 5 | 1023 | Beta-galactosidase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=lacZ PE=1 SV=2 |
| P06864 | 3.72e-147 | 319 | 1347 | 2 | 1004 | Evolved beta-galactosidase subunit alpha OS=Escherichia coli (strain K12) OX=83333 GN=ebgA PE=1 SV=4 |
| Q6LL68 | 1.84e-139 | 364 | 1342 | 44 | 1027 | Beta-galactosidase OS=Photobacterium profundum (strain SS9) OX=298386 GN=lacZ PE=3 SV=1 |
| A6TI29 | 3.06e-139 | 317 | 1338 | 13 | 1019 | Beta-galactosidase 2 OS=Klebsiella pneumoniae subsp. pneumoniae (strain ATCC 700721 / MGH 78578) OX=272620 GN=lacZ2 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
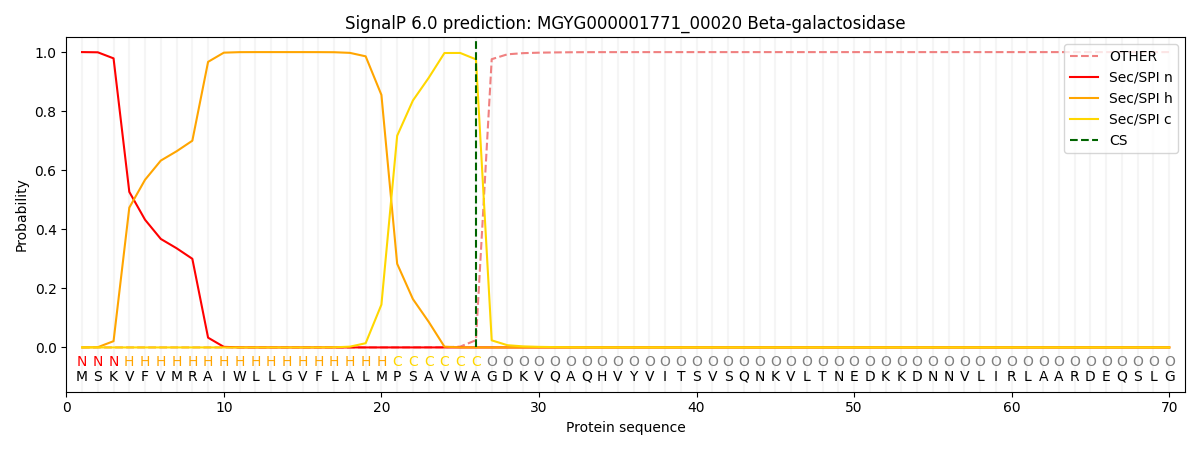
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000311 | 0.999050 | 0.000184 | 0.000152 | 0.000145 | 0.000141 |