You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001963_00078
You are here: Home > Sequence: MGYG000001963_00078
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phocaeicola sp000434735 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola sp000434735 | |||||||||||
| CAZyme ID | MGYG000001963_00078 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 97805; End: 101041 Strand: - | |||||||||||
Full Sequence Download help
| MNNKLKFTLF ALCAPWMAMY AQYNGEPMQK PQEQAKQQQQ QVVQRNKYPH SDNDWENFNV | 60 |
| LHINRLPSAA TFLGYPTREL AREDNKAKSP WFLSLNGTWK FKYVGRYDQR PMDFWKTGFD | 120 |
| LSSWNDIPVP SNWEMEGYGY PFYVGSGYGI KKNPPLIAVE NSPVGSYKRT FTIPADWKGK | 180 |
| QIVVYFGSVA SSFYVWVNGK QVGYSQDSKT PAEFDITPYV KAGENEISVQ VFKFSDGYYL | 240 |
| EDQDFWRLAG IQRDVYVYAR PKVHVRDFEV VTDLDRTYTD ADFHLYVELG KAGGGKISGA | 300 |
| EVEAELQDAQ GKTVYKETKR WNPKEDVMHF MRHVEKPLLW SAEIPNLYRL QIALRVGGKV | 360 |
| QQYITRKIGF RECETRHAQL LVNGKPIYIK GVNRHEHDPY KGHVVDEASM IQDIKLMKEL | 420 |
| NINCVRTSHY PNDPRWYELC DEYGLYVVDE ANIESHGMGY DPNKCLANQP EWEPAFIDRT | 480 |
| ERMFERDKNH PSVIIWSLGN ETGEGCNFAA TYRWVHANDR SQRPVHSEDG IKGPFTDIFC | 540 |
| PMYKKIDVLI NHTLYLPNKP LILCEYAHAM GNSIGNLQDY WDVIEKYPSL QGGHIWDWVD | 600 |
| QGFATKTADG KPFMAYGGDL APKGVPSSAN FCMNGIIAAD RTLKPHAHEV KRVYQNIAFR | 660 |
| LADYHEGLVE LQNKYFFKDL KDFNFYWKLE GNGEELAKGQ IENVALAPQQ KAIFRTSFPK | 720 |
| VTPAPGVEYF VRFYACLKND EGLLKANTEL ADAQAELPFY QPLQAAKQAG KVDMKQTAER | 780 |
| LSLSANGSEV EFDKANGALC SWKQAGKEWI KSPLLPNFWR PVTDNDMGNG MNYTLRAWRE | 840 |
| AGKNARLEEM THHALNEGGY EVVSTYRLDS VAGSTFKITY RMQFRGCLDV ECLFIPANDT | 900 |
| LPLMPRMGVS VGLAREASQM SWLGRGPHEN YCDRNTSAYV GLYHGAVKDE CYLYDRPQEN | 960 |
| GNKTDVRWMS LTDAQGAGLM AIGQPFLSTS AWPYDPASLD EPGLRKSQRH QSDVRESDVI | 1020 |
| TWNLDLKQMG VGGDTSWGAF PHQQYLIPAE RQSFSFRLCP ARSADGAAEG KQYLNMKP | 1078 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 82 | 940 | 5.3e-233 | 0.973404255319149 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 0.0 | 92 | 938 | 12 | 804 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK09525 | lacZ | 0.0 | 53 | 1061 | 15 | 1027 | beta-galactosidase. |
| PRK10340 | ebgA | 0.0 | 53 | 1060 | 2 | 999 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| pfam02836 | Glyco_hydro_2_C | 1.46e-110 | 374 | 659 | 2 | 302 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| smart01038 | Bgal_small_N | 2.58e-108 | 784 | 1058 | 1 | 272 | Beta galactosidase small chain. This domain comprises the small chain of dimeric beta-galactosidases EC:3.2.1.23. This domain is also found in single chain beta-galactosidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ALA72330.1 | 0.0 | 21 | 1077 | 23 | 1076 |
| AII66503.1 | 0.0 | 21 | 1077 | 22 | 1075 |
| AII65460.1 | 0.0 | 21 | 1077 | 22 | 1075 |
| QQY42953.1 | 0.0 | 21 | 1077 | 23 | 1076 |
| QUT56704.1 | 0.0 | 21 | 1077 | 22 | 1075 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6S6Z_A | 5.26e-278 | 54 | 1058 | 3 | 979 | Structureof beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_B Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_C Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_D Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_E Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_F Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_G Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_H Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8] |
| 6SD0_A | 5.43e-278 | 54 | 1058 | 4 | 980 | Structureof beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_B Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_C Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_D Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8] |
| 3DEC_A | 2.01e-233 | 54 | 1062 | 6 | 998 | ChainA, Beta-galactosidase [Bacteroides thetaiotaomicron VPI-5482] |
| 3BGA_A | 5.30e-230 | 54 | 1062 | 9 | 1002 | Crystalstructure of beta-galactosidase from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482],3BGA_B Crystal structure of beta-galactosidase from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482] |
| 3MUY_1 | 4.87e-196 | 54 | 1059 | 15 | 1021 | Chain1, Beta-D-galactosidase [Escherichia coli K-12],3MUY_2 Chain 2, Beta-D-galactosidase [Escherichia coli K-12],3MUY_3 Chain 3, Beta-D-galactosidase [Escherichia coli K-12],3MUY_4 Chain 4, Beta-D-galactosidase [Escherichia coli K-12] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q56307 | 2.97e-277 | 54 | 1058 | 4 | 980 | Beta-galactosidase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=lacZ PE=1 SV=2 |
| O52847 | 3.03e-245 | 46 | 1051 | 15 | 1023 | Beta-galactosidase OS=Priestia megaterium (strain DSM 319 / IMG 1521) OX=592022 GN=bgaM PE=3 SV=1 |
| Q9K9C6 | 5.66e-210 | 50 | 1051 | 4 | 1003 | Beta-galactosidase OS=Alkalihalobacillus halodurans (strain ATCC BAA-125 / DSM 18197 / FERM 7344 / JCM 9153 / C-125) OX=272558 GN=lacZ PE=3 SV=1 |
| A3FEW8 | 1.45e-207 | 54 | 1056 | 17 | 1023 | Beta-galactosidase OS=Enterobacter agglomerans OX=549 GN=lacZ PE=1 SV=2 |
| P81650 | 3.10e-206 | 54 | 1058 | 12 | 1033 | Beta-galactosidase OS=Pseudoalteromonas haloplanktis OX=228 GN=lacZ PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
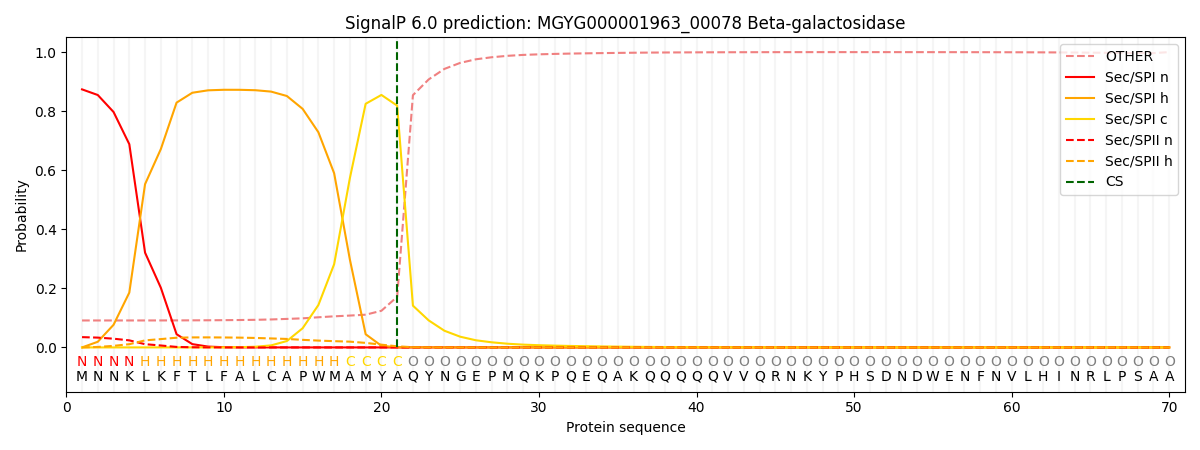
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.106053 | 0.854702 | 0.037685 | 0.000564 | 0.000446 | 0.000524 |
