You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002779_00044
You are here: Home > Sequence: MGYG000002779_00044
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Holdemania sp900120005 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Erysipelotrichales; Erysipelotrichaceae; Holdemania; Holdemania sp900120005 | |||||||||||
| CAZyme ID | MGYG000002779_00044 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 53266; End: 56616 Strand: + | |||||||||||
Full Sequence Download help
| MKKIILGLVI SGLILGGCSS KMDQFVEPQG TPRPSIEPLP TDASFAGEEW YDQMEVWQLH | 60 |
| REPARAALTP YPSSQQALLA ETSALDEIDA ASSSRIQSLN GRWAFYYAAK PTDRLKNLAG | 120 |
| YDAQWYWEDW DTADWDQIDV PSNIQTQWKE DGSFRYEPPI YINQIYPWLN YEAIQYGAQG | 180 |
| QPVAATAVNS VGHYKREFTL DEGLQNKHVF LRFEGVESAF YVYINGQPIG YSEDSYTPAE | 240 |
| FNITPYLKEG VNTIAVEVYR WSDGSYFENQ DFIRLSGIFR DVTLIGREAV EIRDLFVQSD | 300 |
| LAEDYTQAQV QVDVDLRNLS QQPQAGWTLE MELVDQGRAV FDQPLNLESP ELPVLETTEN | 360 |
| QTGTTLSAHF TIDQPKLWTP DQPHLYSLVL TLKNAAGEVM ESVCQRIGLR EIEVQTVDGV | 420 |
| QQMLLNGKPL MLKGVNRHET DPVKGRALGA QEIITDLKLM KAYNINAFRT SHYPNHPLTC | 480 |
| DVADELGLIV VDEANIESHI GEKELGVPGN NPLYNGLIMD RTVSMVERDK NHASVLFWSL | 540 |
| GNEATYREYE MNENYPFYNT SMWILQRDPR RLRVYERDNR IGNTRETSMV DVASSQYWTL | 600 |
| DQLKDYAKKN KHAFFQSEYI HAMGNGVANL AEYFALFKTY PQLQGGFIWD FIDQTILTGD | 660 |
| PVTGQFYYGY GSDWGTPLND GDFCGNGLVN ADRTPSAELE EVKKVQQDVS FTYDSKKRRL | 720 |
| TITNEFLATD LNAFAVELNF YQNGTLFHTA ALSEAEKALP PGMSKTIDVA LPDYDSDAEV | 780 |
| ILEIDVKYSQ DQSWANAYGG KQGDILAFEQ FVLQEGKPAA FVSPDLSLTV SEQEGKWTIQ | 840 |
| GRTEQNQDFE MILNPAKMEI EAYRLGDQTF LTGGPQPSFY RAETSGDPQF SEAVRQAGKA | 900 |
| VEYKSPSIQR KEENGEVKQL TIKASGTINE FNTSLETEIT IQGNGQVAVA MTMKVPSKEK | 960 |
| VGPLARAGLT LSLDASLNEI RYNGRGPEEN YIDRHTGSRL GQWTAKINDF YNDQLLKPQD | 1020 |
| SGNRTEVRSL TLNGETNALT LTFDEPVGMN IMTWDELSLA SAAHMKDAQK LDQPLLHLDA | 1080 |
| AQRGLGNGSW GAEPLKEYQI KQNQTYTVSF QIKPDQ | 1116 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 87 | 1000 | 3.2e-185 | 0.9720744680851063 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK09525 | lacZ | 0.0 | 56 | 1113 | 22 | 1027 | beta-galactosidase. |
| PRK10340 | ebgA | 0.0 | 54 | 1116 | 1 | 1001 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| COG3250 | LacZ | 1.62e-139 | 93 | 1002 | 9 | 808 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| pfam02836 | Glyco_hydro_2_C | 5.60e-85 | 422 | 711 | 7 | 302 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| pfam02929 | Bgal_small_N | 1.04e-51 | 845 | 1112 | 1 | 230 | Beta galactosidase small chain. This domain comprises the small chain of dimeric beta-galactosidases EC:3.2.1.23. This domain is also found in single chain beta-galactosidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| APC49533.1 | 4.03e-202 | 49 | 1114 | 35 | 1044 |
| QKY70370.1 | 1.98e-200 | 49 | 1114 | 37 | 1046 |
| QYR22672.1 | 7.52e-199 | 49 | 1113 | 16 | 1035 |
| QZN78339.1 | 2.41e-193 | 49 | 1114 | 16 | 1038 |
| AKO22786.1 | 1.44e-191 | 49 | 1114 | 16 | 1038 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6S6Z_A | 8.28e-164 | 98 | 1113 | 40 | 980 | Structureof beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_B Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_C Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_D Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_E Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_F Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_G Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_H Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8] |
| 6SD0_A | 8.49e-164 | 98 | 1113 | 41 | 981 | Structureof beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_B Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_C Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_D Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8] |
| 4V41_A | 6.43e-120 | 93 | 1100 | 48 | 1009 | E.COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) [Escherichia coli BL21],4V41_B E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) [Escherichia coli BL21],4V41_C E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) [Escherichia coli BL21],4V41_D E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) [Escherichia coli BL21],4V41_E E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) [Escherichia coli BL21],4V41_F E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) [Escherichia coli BL21],4V41_G E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) [Escherichia coli BL21],4V41_H E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) [Escherichia coli BL21],4V41_I E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) [Escherichia coli BL21],4V41_J E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) [Escherichia coli BL21],4V41_K E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) [Escherichia coli BL21],4V41_L E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) [Escherichia coli BL21],4V41_M E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) [Escherichia coli BL21],4V41_N E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) [Escherichia coli BL21],4V41_O E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) [Escherichia coli BL21],4V41_P E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) [Escherichia coli BL21],4V44_A E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH 2-F-LACTOSE [Escherichia coli],4V44_B E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH 2-F-LACTOSE [Escherichia coli],4V44_C E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH 2-F-LACTOSE [Escherichia coli],4V44_D E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH 2-F-LACTOSE [Escherichia coli],4V44_E E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH 2-F-LACTOSE [Escherichia coli],4V44_F E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH 2-F-LACTOSE [Escherichia coli],4V44_G E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH 2-F-LACTOSE [Escherichia coli],4V44_H E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH 2-F-LACTOSE [Escherichia coli],4V44_I E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH 2-F-LACTOSE [Escherichia coli],4V44_J E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH 2-F-LACTOSE [Escherichia coli],4V44_K E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH 2-F-LACTOSE [Escherichia coli],4V44_L E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH 2-F-LACTOSE [Escherichia coli],4V44_M E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH 2-F-LACTOSE [Escherichia coli],4V44_N E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH 2-F-LACTOSE [Escherichia coli],4V44_O E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH 2-F-LACTOSE [Escherichia coli],4V44_P E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH 2-F-LACTOSE [Escherichia coli],4V45_A E. COLI (lacZ) BETA-GALACTOSIDASE-TRAPPED 2-F-GALACTOSYL-ENZYME INTERMEDIATE [Escherichia coli],4V45_B E. COLI (lacZ) BETA-GALACTOSIDASE-TRAPPED 2-F-GALACTOSYL-ENZYME INTERMEDIATE [Escherichia coli],4V45_C E. COLI (lacZ) BETA-GALACTOSIDASE-TRAPPED 2-F-GALACTOSYL-ENZYME INTERMEDIATE [Escherichia coli],4V45_D E. COLI (lacZ) BETA-GALACTOSIDASE-TRAPPED 2-F-GALACTOSYL-ENZYME INTERMEDIATE [Escherichia coli],4V45_E E. COLI (lacZ) BETA-GALACTOSIDASE-TRAPPED 2-F-GALACTOSYL-ENZYME INTERMEDIATE [Escherichia coli],4V45_F E. COLI (lacZ) BETA-GALACTOSIDASE-TRAPPED 2-F-GALACTOSYL-ENZYME INTERMEDIATE [Escherichia coli],4V45_G E. COLI (lacZ) BETA-GALACTOSIDASE-TRAPPED 2-F-GALACTOSYL-ENZYME INTERMEDIATE [Escherichia coli],4V45_H E. COLI (lacZ) BETA-GALACTOSIDASE-TRAPPED 2-F-GALACTOSYL-ENZYME INTERMEDIATE [Escherichia coli],4V45_I E. COLI (lacZ) BETA-GALACTOSIDASE-TRAPPED 2-F-GALACTOSYL-ENZYME INTERMEDIATE [Escherichia coli],4V45_J E. COLI (lacZ) BETA-GALACTOSIDASE-TRAPPED 2-F-GALACTOSYL-ENZYME INTERMEDIATE [Escherichia coli],4V45_K E. COLI (lacZ) BETA-GALACTOSIDASE-TRAPPED 2-F-GALACTOSYL-ENZYME INTERMEDIATE [Escherichia coli],4V45_L E. COLI (lacZ) BETA-GALACTOSIDASE-TRAPPED 2-F-GALACTOSYL-ENZYME INTERMEDIATE [Escherichia coli],4V45_M E. COLI (lacZ) BETA-GALACTOSIDASE-TRAPPED 2-F-GALACTOSYL-ENZYME INTERMEDIATE [Escherichia coli],4V45_N E. COLI (lacZ) BETA-GALACTOSIDASE-TRAPPED 2-F-GALACTOSYL-ENZYME INTERMEDIATE [Escherichia coli],4V45_O E. COLI (lacZ) BETA-GALACTOSIDASE-TRAPPED 2-F-GALACTOSYL-ENZYME INTERMEDIATE [Escherichia coli],4V45_P E. COLI (lacZ) BETA-GALACTOSIDASE-TRAPPED 2-F-GALACTOSYL-ENZYME INTERMEDIATE [Escherichia coli] |
| 1JZ7_A | 1.72e-119 | 93 | 1100 | 48 | 1009 | E.COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH GALACTOSE [Escherichia coli],1JZ7_B E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH GALACTOSE [Escherichia coli],1JZ7_C E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH GALACTOSE [Escherichia coli],1JZ7_D E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH GALACTOSE [Escherichia coli],4TTG_A Beta-galactosidase (E. coli) in the presence of potassium chloride. [Escherichia coli],4TTG_B Beta-galactosidase (E. coli) in the presence of potassium chloride. [Escherichia coli],4TTG_C Beta-galactosidase (E. coli) in the presence of potassium chloride. [Escherichia coli],4TTG_D Beta-galactosidase (E. coli) in the presence of potassium chloride. [Escherichia coli] |
| 3MUY_1 | 1.72e-119 | 93 | 1100 | 48 | 1009 | Chain1, Beta-D-galactosidase [Escherichia coli K-12],3MUY_2 Chain 2, Beta-D-galactosidase [Escherichia coli K-12],3MUY_3 Chain 3, Beta-D-galactosidase [Escherichia coli K-12],3MUY_4 Chain 4, Beta-D-galactosidase [Escherichia coli K-12] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O52847 | 3.11e-185 | 49 | 1114 | 19 | 1033 | Beta-galactosidase OS=Priestia megaterium (strain DSM 319 / IMG 1521) OX=592022 GN=bgaM PE=3 SV=1 |
| Q56307 | 4.65e-163 | 98 | 1113 | 41 | 981 | Beta-galactosidase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=lacZ PE=1 SV=2 |
| Q9K9C6 | 1.86e-153 | 50 | 1112 | 9 | 1011 | Beta-galactosidase OS=Alkalihalobacillus halodurans (strain ATCC BAA-125 / DSM 18197 / FERM 7344 / JCM 9153 / C-125) OX=272558 GN=lacZ PE=3 SV=1 |
| A5F5U6 | 1.19e-135 | 97 | 1113 | 48 | 1019 | Beta-galactosidase OS=Vibrio cholerae serotype O1 (strain ATCC 39541 / Classical Ogawa 395 / O395) OX=345073 GN=lacZ PE=3 SV=2 |
| P24131 | 8.09e-135 | 49 | 1001 | 9 | 891 | Beta-galactosidase OS=Clostridium acetobutylicum OX=1488 GN=cbgA PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
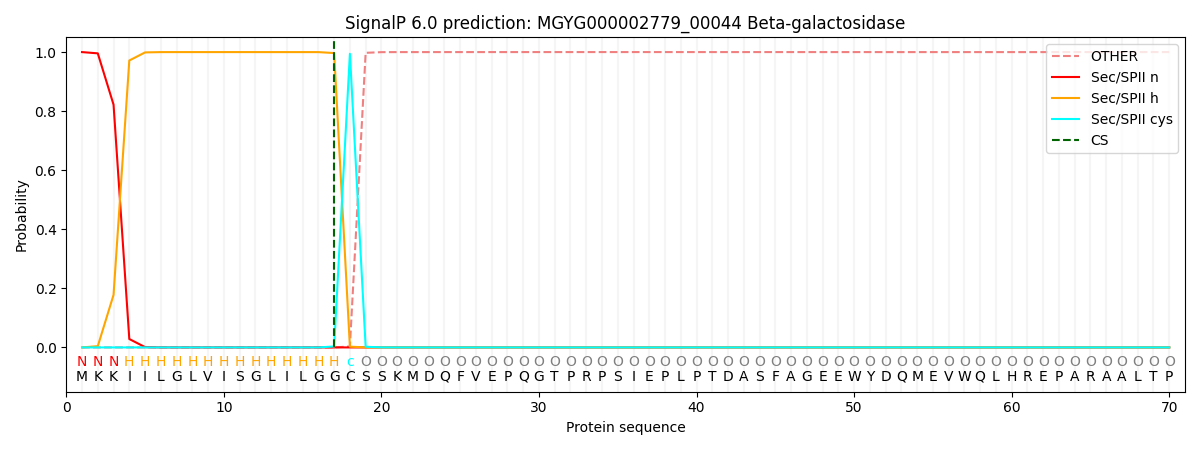
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000053 | 0.000000 | 0.000000 | 0.000000 |
