You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003019_00235
You are here: Home > Sequence: MGYG000003019_00235
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes sp900553175 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; Alistipes sp900553175 | |||||||||||
| CAZyme ID | MGYG000003019_00235 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 58483; End: 61566 Strand: - | |||||||||||
Full Sequence Download help
| MKRTLHLLAA LLLTSAAAAA PTIPEWQDPT VNAINRLPMR ATFAAGERLS LDGIWKFRWV | 60 |
| RNAWERPEDF GRPDCDDRAW GTIPVPGMWE MNGYGDPVYV NTGYAWRGSF RSEPPRVPEA | 120 |
| ENHVGSYRRT FELPASWSGK DIFLHIGSAT SNVCVWINGR FVGYSEDSKL AASFEVTEFV | 180 |
| RPGRNLIALQ IFRWCDGTYL EDQDFWRMSG IARGVELTAR DRAHLCDLHV TPALDDDHAV | 240 |
| GRLQVEMTAT PRVKELRLEL RDAAGQTVAR AAAAPVSGQT GCSLEVENPA LWSAEEPNLY | 300 |
| TLSVEVVDRR DRTAETAETA VGFRTVEIRG GRLLVNGAPV LIKGVNRHEM DPLGGYVLSR | 360 |
| ERMMEDIRIM KELNINAVRT CHYPDDPFWY DLCDRYGIYV FDEADVESHG MGYGERTLAR | 420 |
| DPAYARAHLE RNSRMVARDR NHPSVIVWSM GNEAGMGPNF EACYEWIKAH DPSRPVHYER | 480 |
| AFGYRRASDG KEFTDIACPM YHDYAWCRRY CESEPDKPLI QCEYAHAMGN SLGGFDTYWE | 540 |
| LIRRYPCYQG GFIWDFADQG LARYESDGRA SFLYGGDFNG SDPSDNSFNC NGIVAADRTW | 600 |
| HPHAREVQRQ YQNIWTSLAD PARGLVEVFN ENFFSTLDAY ELEWQLLQDG LVTRRGRVET | 660 |
| CDIGPRQRRT VALGFSEADF GNEGREVLLN VAYNLRQPAG LLPAGLLPAG FTAARQQLTI | 720 |
| RTAGRTAGFG PASKGSVAVV RRERAVCAEG DGWHILFGRD GWVKGYCVRG QELLLEGSSL | 780 |
| RPQFWRAPTE NDLGNGTARR SAVWKKPLMK LLSFDASAET EGMALVEARY ELPEVGAQLT | 840 |
| MTYRIDGAGA MTVTERIAPG DGAPAPDLLR FGMRFEMPAR YDRIVCYGRG PHENYCDRLT | 900 |
| SADLGIWQQR VADQYHDRYV RPQESGTRSD LRWWRVEDSA GAGISIAADA PFSASALPYL | 960 |
| TEDLDVTELP PQQHSGTLVP RHATCVNIEA RQAGLGCIDS WGALPEEEHR LPYGEYTFRF | 1020 |
| VLRPGQK | 1027 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 44 | 902 | 1.2e-216 | 0.9627659574468085 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK09525 | lacZ | 0.0 | 26 | 1024 | 17 | 1026 | beta-galactosidase. |
| PRK10340 | ebgA | 0.0 | 26 | 1026 | 4 | 1001 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| COG3250 | LacZ | 4.32e-156 | 47 | 906 | 11 | 808 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| pfam02836 | Glyco_hydro_2_C | 1.67e-108 | 326 | 614 | 1 | 300 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| smart01038 | Bgal_small_N | 1.54e-90 | 749 | 1022 | 1 | 272 | Beta galactosidase small chain. This domain comprises the small chain of dimeric beta-galactosidases EC:3.2.1.23. This domain is also found in single chain beta-galactosidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBL01806.1 | 0.0 | 1 | 1027 | 2 | 1018 |
| BBL09676.1 | 0.0 | 1 | 1027 | 2 | 1018 |
| BBL12470.1 | 0.0 | 1 | 1027 | 2 | 1018 |
| AFL76749.1 | 0.0 | 1 | 1027 | 1 | 1017 |
| BBL06016.1 | 0.0 | 1 | 1027 | 1 | 1022 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6S6Z_A | 1.25e-213 | 47 | 1027 | 37 | 984 | Structureof beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_B Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_C Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_D Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_E Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_F Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_G Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_H Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8] |
| 6SD0_A | 1.28e-213 | 47 | 1027 | 38 | 985 | Structureof beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_B Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_C Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_D Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8] |
| 3DEC_A | 1.28e-209 | 24 | 1024 | 5 | 996 | ChainA, Beta-galactosidase [Bacteroides thetaiotaomicron VPI-5482] |
| 3BGA_A | 6.96e-198 | 23 | 1024 | 7 | 1000 | Crystalstructure of beta-galactosidase from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482],3BGA_B Crystal structure of beta-galactosidase from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482] |
| 6CVM_A | 2.12e-184 | 12 | 1021 | 1 | 1018 | Atomicresolution cryo-EM structure of beta-galactosidase [Escherichia coli K-12],6CVM_B Atomic resolution cryo-EM structure of beta-galactosidase [Escherichia coli K-12],6CVM_C Atomic resolution cryo-EM structure of beta-galactosidase [Escherichia coli K-12],6CVM_D Atomic resolution cryo-EM structure of beta-galactosidase [Escherichia coli K-12],6X1Q_A 1.8 Angstrom resolution structure of b-galactosidase with a 200 kV cryoARM electron microscope [Escherichia coli K-12],6X1Q_B 1.8 Angstrom resolution structure of b-galactosidase with a 200 kV cryoARM electron microscope [Escherichia coli K-12],6X1Q_C 1.8 Angstrom resolution structure of b-galactosidase with a 200 kV cryoARM electron microscope [Escherichia coli K-12],6X1Q_D 1.8 Angstrom resolution structure of b-galactosidase with a 200 kV cryoARM electron microscope [Escherichia coli K-12] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q56307 | 7.03e-213 | 47 | 1027 | 38 | 985 | Beta-galactosidase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=lacZ PE=1 SV=2 |
| O52847 | 6.72e-202 | 18 | 1024 | 12 | 1033 | Beta-galactosidase OS=Priestia megaterium (strain DSM 319 / IMG 1521) OX=592022 GN=bgaM PE=3 SV=1 |
| Q9K9C6 | 9.57e-188 | 27 | 1011 | 11 | 999 | Beta-galactosidase OS=Alkalihalobacillus halodurans (strain ATCC BAA-125 / DSM 18197 / FERM 7344 / JCM 9153 / C-125) OX=272558 GN=lacZ PE=3 SV=1 |
| Q3Z583 | 1.16e-184 | 11 | 1021 | 2 | 1020 | Beta-galactosidase OS=Shigella sonnei (strain Ss046) OX=300269 GN=lacZ PE=3 SV=1 |
| A0KQH4 | 1.68e-184 | 25 | 1023 | 10 | 1025 | Beta-galactosidase OS=Aeromonas hydrophila subsp. hydrophila (strain ATCC 7966 / DSM 30187 / BCRC 13018 / CCUG 14551 / JCM 1027 / KCTC 2358 / NCIMB 9240 / NCTC 8049) OX=380703 GN=lacZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
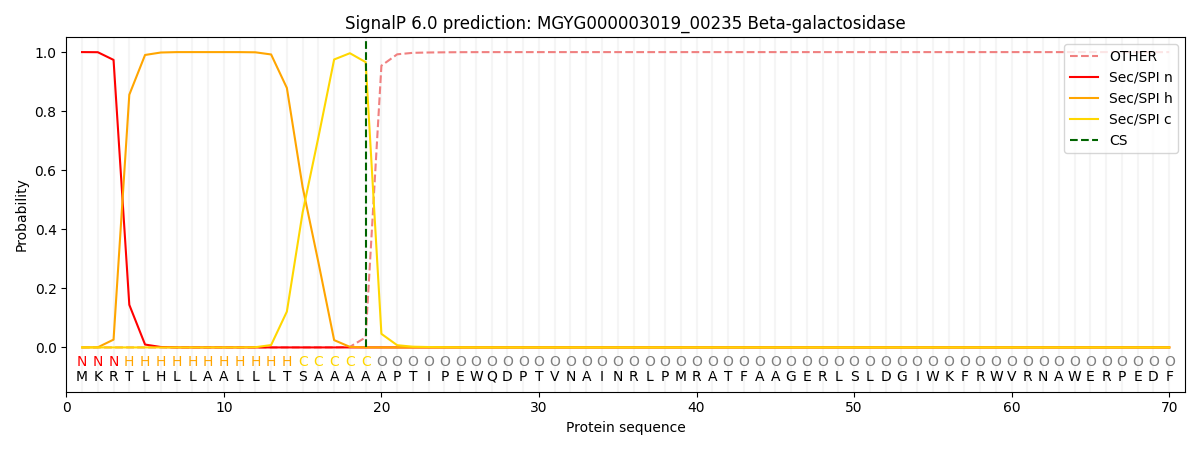
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000428 | 0.998816 | 0.000200 | 0.000198 | 0.000176 | 0.000164 |
