You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003193_00052
You are here: Home > Sequence: MGYG000003193_00052
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-831 sp900544175 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA932; CAG-831; CAG-831 sp900544175 | |||||||||||
| CAZyme ID | MGYG000003193_00052 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 56545; End: 59982 Strand: + | |||||||||||
Full Sequence Download help
| MKNILIPLAL VLCPAVLSAQ GQKDRLPYWQ DVQTVEVNRE YPRTAFMSYP DSATAAAYRY | 60 |
| ENSPRYLLLN GVWKFYFVDA YRDLPENIAD PSVDASGWND INVPGNWEVQ GFGTAIYTNH | 120 |
| GYEFKPRNPT PPVLPEDNPV GVYRREFTVP EAWDGSEIFL NVDGAKSGVY VYLNGREVGY | 180 |
| SEDSKTSAEF AVSDFLVPGV NTLVLKIYRW STGSYLECQD FWRMSGIERD VYLWCQPREA | 240 |
| ALKDFTVVST LDDTYTEGLF SLRAVLKNTS ASGSEAVLRG TLTGPDGAVV WSDVRNVPLD | 300 |
| AGSTSEVLFE NRIPDVQPWS AEHPDLYRLT MTVYADGKAV ETVPYHVGFR KFEIKPSGEL | 360 |
| SPEGRPYVLF YVNGQPVKLK GVNTHEHNPM TGHYVTEELI RRDLELMKQN NINTVRLAHY | 420 |
| PQGRRFYELC DEYGLYVYDE ANIESHGMYY NLRKGGTLGN NPEWLAAHMD RTVNMYERNK | 480 |
| NYPCITVWSL GNEAGNGYNF YQTYLWIKDR ERGGMDRPVC YERAQWEWNS DMYVPQYPGA | 540 |
| DWLRQIGRSG SDRPVVPSEY SHAMGNSSGS LSLQWDEIYR YPNLQGGYIW DWVDQGIWQD | 600 |
| RDGGFWAYGG DFGVNAPSDG NFLCNGIVNP DRNPHPAMTE VKHAYQNAAF RPSGYAEDPT | 660 |
| AERTLATGRL AFSSLPSGGK VSVDVINRFY FTDLDAYDFT YELTADGRVA ASGTLAVSAA | 720 |
| PQDTVTVEIP VPALKSSGTE YFLNLYMKQR SEVLGIPAGH VLASDQIAFP VTGERPVYAE | 780 |
| GRGPALSVDS VSSEGVISIF SPSVEWVFDR SAAAVTSYKV RGVEYFAGGF GLRPNFWRGP | 840 |
| NDNDYGNGAP KRLQIWKTSS QELRVGAVSV AMDGADAVMT VEYLLAAGNK YIAEYRVHPD | 900 |
| GVVDARLHFT PALMPEVESE ASEATLTATY TPGQESRVGL KARPEVPRIG VRFRLPVAMH | 960 |
| GVTWYGRGPG ENYIDRSRGS LVGLYSSSAE AMYFPYVRPQ ENGHRSDTRW AAFYGGGNGL | 1020 |
| MVVADSTIGF SALRNPVEDF DSEEAVEHDY QWPNFTPEQI ASKDPADAKD VLRRMHHIND | 1080 |
| IVPQDYVEVC IDMRQQGVAG YDSWGDRPEP QYQLPADRDY TWSFTLVPVK NARDAQRKAL | 1140 |
| YDYGL | 1145 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 57 | 982 | 2.8e-201 | 0.973404255319149 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK09525 | lacZ | 0.0 | 29 | 1126 | 17 | 1024 | beta-galactosidase. |
| PRK10340 | ebgA | 0.0 | 29 | 1145 | 4 | 1017 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| COG3250 | LacZ | 4.78e-131 | 69 | 976 | 15 | 800 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| smart01038 | Bgal_small_N | 9.11e-90 | 801 | 1126 | 2 | 272 | Beta galactosidase small chain. This domain comprises the small chain of dimeric beta-galactosidases EC:3.2.1.23. This domain is also found in single chain beta-galactosidase. |
| pfam02836 | Glyco_hydro_2_C | 2.33e-87 | 369 | 650 | 7 | 302 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUR46524.1 | 0.0 | 21 | 1143 | 2 | 1101 |
| QUT33020.1 | 0.0 | 18 | 1143 | 10 | 1112 |
| QRN02091.1 | 0.0 | 21 | 1143 | 2 | 1101 |
| QDH57581.1 | 0.0 | 21 | 1143 | 2 | 1101 |
| QNL36573.1 | 0.0 | 17 | 1143 | 23 | 1126 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6SD0_A | 4.26e-212 | 26 | 1126 | 1 | 980 | Structureof beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_B Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_C Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_D Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8] |
| 6S6Z_A | 8.19e-212 | 27 | 1126 | 1 | 979 | Structureof beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_B Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_C Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_D Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_E Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_F Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_G Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_H Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8] |
| 3DEC_A | 3.25e-203 | 27 | 1130 | 5 | 998 | ChainA, Beta-galactosidase [Bacteroides thetaiotaomicron VPI-5482] |
| 3BGA_A | 1.22e-198 | 23 | 1130 | 4 | 1002 | Crystalstructure of beta-galactosidase from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482],3BGA_B Crystal structure of beta-galactosidase from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482] |
| 3MUY_1 | 1.79e-142 | 29 | 1009 | 16 | 942 | Chain1, Beta-D-galactosidase [Escherichia coli K-12],3MUY_2 Chain 2, Beta-D-galactosidase [Escherichia coli K-12],3MUY_3 Chain 3, Beta-D-galactosidase [Escherichia coli K-12],3MUY_4 Chain 4, Beta-D-galactosidase [Escherichia coli K-12] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q56307 | 2.33e-211 | 26 | 1126 | 1 | 980 | Beta-galactosidase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=lacZ PE=1 SV=2 |
| O52847 | 1.39e-188 | 27 | 1129 | 18 | 1034 | Beta-galactosidase OS=Priestia megaterium (strain DSM 319 / IMG 1521) OX=592022 GN=bgaM PE=3 SV=1 |
| Q6LL68 | 7.44e-167 | 29 | 1131 | 12 | 1030 | Beta-galactosidase OS=Photobacterium profundum (strain SS9) OX=298386 GN=lacZ PE=3 SV=1 |
| A5F5U6 | 1.40e-164 | 29 | 1126 | 13 | 1016 | Beta-galactosidase OS=Vibrio cholerae serotype O1 (strain ATCC 39541 / Classical Ogawa 395 / O395) OX=345073 GN=lacZ PE=3 SV=2 |
| B4S2K9 | 1.20e-156 | 29 | 1125 | 13 | 1036 | Beta-galactosidase OS=Alteromonas mediterranea (strain DSM 17117 / CIP 110805 / LMG 28347 / Deep ecotype) OX=1774373 GN=lacZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
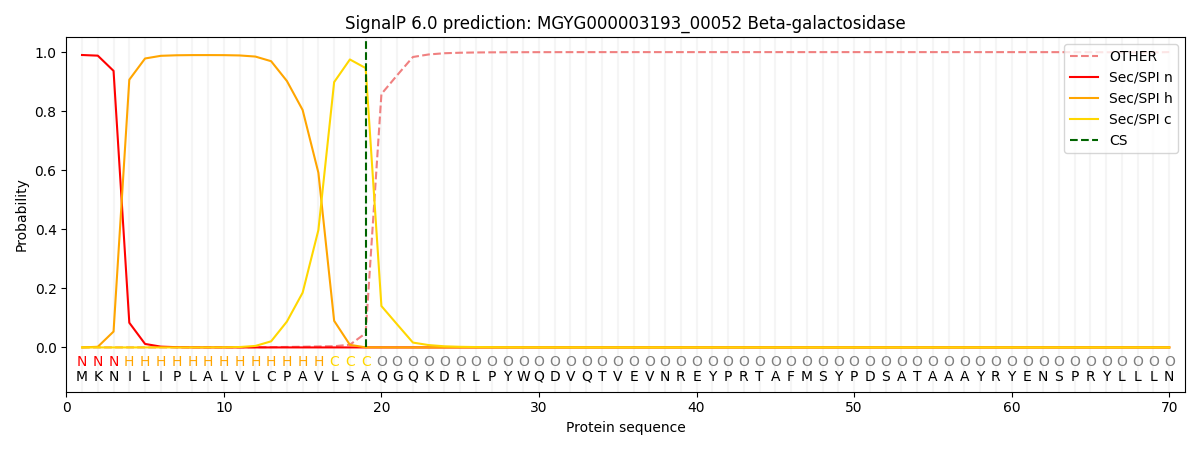
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001248 | 0.986747 | 0.011118 | 0.000331 | 0.000275 | 0.000259 |
