You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003535_00028
You are here: Home > Sequence: MGYG000003535_00028
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
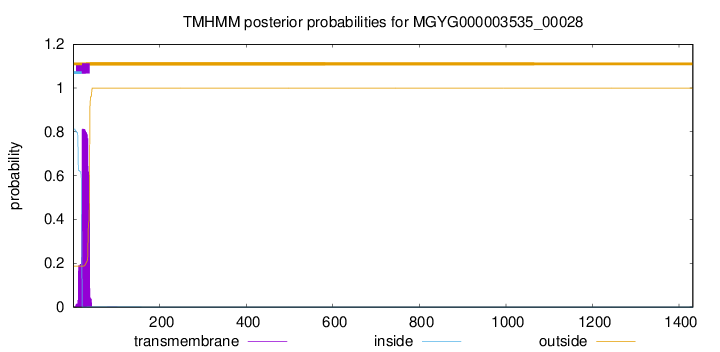
TMHMM annotations
Basic Information help
| Species | Prevotella sp004556065 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp004556065 | |||||||||||
| CAZyme ID | MGYG000003535_00028 | |||||||||||
| CAZy Family | GH137 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 12794; End: 17092 Strand: - | |||||||||||
Full Sequence Download help
| MRQSSLLTNP IIHAYIMKNT LLLSLLMISL TTLAQTVTDD VMQRIYDEVK SPYKYGMVIA | 60 |
| PPDNGHKIDC PTVYREGGKW YMTYVIYNGS GGLDGRGYTT WLSESDDLLH WRQLGCILDY | 120 |
| KDSGWDMNQR GGFPSLINWE WGGDYRMSAY KGRHWMTYIG GEGTGYEGVR APLYIGHAWT | 180 |
| KAGNGALTTA HEWKSGDKPL MHVDDKDAQW WEKLIQYKST IYEDKRRTLG ARFVMFYNAG | 240 |
| GINPDNNHKA ERIGIALSDN MRKWRRYAGN PVFAHEVPGI ITGDAQIVDF TRCNAVQGVS | 300 |
| EEPLYVMFYF SAFNPKRKYS AYNTFAVSRD LVNWKEWEGA DLVYPTKDYE NLFAHKSCVV | 360 |
| KHNGVVYHFY CAVNTAEQRG IALATSKPMG KSPVTFVAPD KKGKRDIISL NDGWKTTLYD | 420 |
| NGKAVETKTV NVPHNWDDYY GYRQLKHGNL HGTATYCRTL NIDKQTEKRY FLQLEGAGSY | 480 |
| ATVTVNGKQF DRRPVGRTTL TLDITDALRG GKNNIEVRCD HPELITDMPW VCGGCSSEWG | 540 |
| FSEGSQPLGL FRPVSLVVTD NVRIEPFGVH VWNNAACDSV FIETEVRNYS AETVSAEIVN | 600 |
| SFNLASGKKA FRLNDTITLS PGETRVIRQS SPIANVTRWS PSDPYLYNLV SMVKRSGKTT | 660 |
| DQLTTPYGIR DIKWVREGAA PGFFINGQRL FINGTCEYEH LLGGGHAFSP EQIKARMRLI | 720 |
| RQAGFNAYRE AHQPHNLLYQ SLLDREGMLF WSQFSAHIWY DTPQFRENFK MLMRQWIKER | 780 |
| RNSPSIILWG LQNESTLPRD FAEECTAIIR EMDPTCASQP GLAGRLVTTC NGGEGSDWNV | 840 |
| IQNWSGTYGG TALNYDKELK QPSQLLNGEY GAWRTLGLHT EAPIDSIRVS KSYTEERATD | 900 |
| LLETKVRLAE EAADSVCGHF QWIFATHDNP GRVQPDGALR MADKIGPVNF KGLTTVWEQP | 960 |
| TDAFFMYRSN YVSPDTDPMV YIASHTWADR FKTSGPRRTD IAVYSNCDSV MLYNSADNSV | 1020 |
| FLGRKRNARK PGTHMLWEHR KVEYNVLRAV GYRHGKPVAE DIILLEGLAE APEFDSLYGP | 1080 |
| SAVVPQAADN NRDILKPETG AYCVRLNCGG NAYTDSYGNR WEADDTSFSH SWGETFASSR | 1140 |
| DDIPQLVRAS QSSVAVPVHG TRDWQLFQTF RYGRHRLAFD FPLPDGEYAV EMYFAEPWLG | 1200 |
| IGGGVKTDCE GQRLFSVAIN GTVRIADLDI WAEAGYAGAL KKTVIAEAKD GHLRISFPET | 1260 |
| KAGQAVISAI AVRQILPEGT SQAAAVAPLR AKKDNFWDAI NADKVERLPK NLLPADDEAF | 1320 |
| PATRYTPVTG LRTKDKAETM FRITPGVARE YALRFRYKNV GEARTARMRI IDEKGSILVD | 1380 |
| RDILFPTTPK KFKLVSTTTG TQINAGNYHL QLLQPGTTKS LKGVEFEYLE VQ | 1432 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH137 | 35 | 396 | 1.1e-137 | 0.9911764705882353 |
| GH2 | 400 | 857 | 7.3e-82 | 0.5186170212765957 |
| CBM57 | 1104 | 1270 | 1.2e-42 | 0.9863945578231292 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 8.44e-37 | 404 | 795 | 9 | 407 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| pfam11721 | Malectin | 8.62e-35 | 1102 | 1270 | 1 | 164 | Di-glucose binding within endoplasmic reticulum. Malectin is a membrane-anchored protein of the endoplasmic reticulum that recognizes and binds Glc2-N-glycan. It carries a signal peptide from residues 1-26, a C-terminal transmembrane helix from residues 255-274, and a highly conserved central part of approximately 190 residues followed by an acidic, glutamate-rich region. Carbohydrate-binding is mediated by the four aromatic residues, Y67, Y89, Y116, and F117 and the aspartate at D186. NMR-based ligand-screening studies has shown binding of the protein to maltose and related oligosaccharides, on the basis of which the protein has been designated "malectin", and its endogenous ligand is found to be Glc2-high-mannose N-glycan. |
| PRK10340 | ebgA | 5.41e-26 | 432 | 815 | 76 | 469 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| PRK10150 | PRK10150 | 1.42e-16 | 466 | 925 | 80 | 556 | beta-D-glucuronidase; Provisional |
| pfam02837 | Glyco_hydro_2_N | 7.72e-14 | 409 | 559 | 3 | 168 | Glycosyl hydrolases family 2, sugar binding domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities and has a jelly-roll fold. The domain binds the sugar moiety during the sugar-hydrolysis reaction. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT45339.1 | 0.0 | 37 | 1432 | 26 | 1427 |
| QRQ48836.1 | 0.0 | 37 | 1432 | 26 | 1427 |
| QQY43469.1 | 0.0 | 17 | 1432 | 1 | 1426 |
| ALK84974.1 | 0.0 | 17 | 1432 | 1 | 1434 |
| AII68813.1 | 0.0 | 17 | 1432 | 1 | 1423 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5MUI_A | 7.90e-172 | 37 | 402 | 29 | 380 | Glycosidehydrolase BT_0996 [Bacteroides thetaiotaomicron VPI-5482] |
| 5MT2_A | 5.83e-166 | 37 | 402 | 29 | 380 | Glycosidehydrolase BT_0996 [Bacteroides thetaiotaomicron VPI-5482],5MUJ_A BT0996 RGII Chain B Complex [Bacteroides thetaiotaomicron VPI-5482] |
| 6D50_A | 2.17e-50 | 396 | 1275 | 24 | 885 | Bacteroidesuniforms beta-glucuronidase 2 bound to D-glucaro-1,5-lactone [Bacteroides uniformis str. 3978 T3 ii],6D50_B Bacteroides uniforms beta-glucuronidase 2 bound to D-glucaro-1,5-lactone [Bacteroides uniformis str. 3978 T3 ii] |
| 6D8G_A | 5.11e-50 | 396 | 1275 | 24 | 885 | D341AD367A calcium binding mutant of Bacteroides uniformis beta-glucuronidase 2 [Bacteroides uniformis str. 3978 T3 ii],6D8G_B D341A D367A calcium binding mutant of Bacteroides uniformis beta-glucuronidase 2 [Bacteroides uniformis str. 3978 T3 ii] |
| 5UJ6_A | 8.27e-49 | 404 | 1275 | 24 | 877 | CrystalStructure of Bacteroides Uniformis beta-glucuronidase [Bacteroides uniformis str. 3978 T3 ii],5UJ6_B Crystal Structure of Bacteroides Uniformis beta-glucuronidase [Bacteroides uniformis str. 3978 T3 ii],6NZG_A Bacteroides uniformis beta-glucuronidase 2 covalently bound to cyclophellitol-6-carboxylate aziridine [Bacteroides uniformis],6NZG_B Bacteroides uniformis beta-glucuronidase 2 covalently bound to cyclophellitol-6-carboxylate aziridine [Bacteroides uniformis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KPJ7 | 2.78e-45 | 428 | 1064 | 80 | 718 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_21970 PE=2 SV=1 |
| T2KM09 | 2.20e-34 | 409 | 1058 | 50 | 700 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22050 PE=2 SV=2 |
| P26257 | 1.72e-19 | 405 | 1012 | 2 | 583 | Beta-galactosidase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=lacZ PE=1 SV=1 |
| A9MQ82 | 2.87e-18 | 407 | 795 | 52 | 463 | Beta-galactosidase OS=Salmonella arizonae (strain ATCC BAA-731 / CDC346-86 / RSK2980) OX=41514 GN=lacZ PE=3 SV=2 |
| P06864 | 2.88e-18 | 453 | 795 | 110 | 450 | Evolved beta-galactosidase subunit alpha OS=Escherichia coli (strain K12) OX=83333 GN=ebgA PE=1 SV=4 |
SignalP and Lipop Annotations help
This protein is predicted as SP
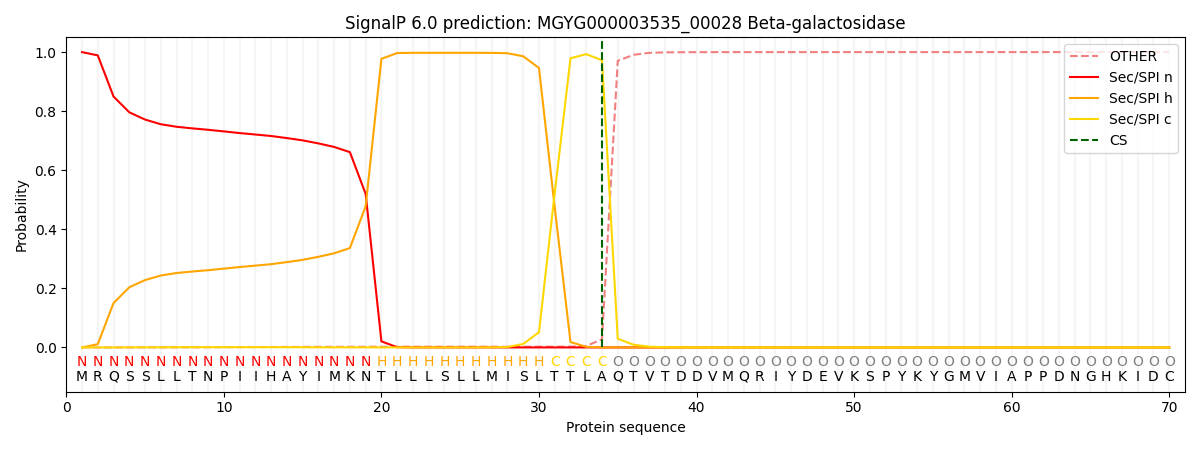
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000395 | 0.998937 | 0.000171 | 0.000167 | 0.000151 | 0.000144 |