You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003746_00149
You are here: Home > Sequence: MGYG000003746_00149
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Porphyromonadaceae; Porphyromonas_A; | |||||||||||
| CAZyme ID | MGYG000003746_00149 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 4958; End: 8941 Strand: - | |||||||||||
Full Sequence Download help
| MKRLFLPLLV LLTLSLPGQG QKLHPIAGYE YGTAVAPDGS EWESPERLGL NREQPAAYFF | 60 |
| SYRSVEAARQ YLPETPGTYY RSLDGAWRFH WVGNPGERPM DFYRPGYDVS GWDEISVPSC | 120 |
| WSVAGIQPDG SLKYGTPIYC NQPYIFKHTV APGDWKGGVM RTPPTDWTTY KDRNEVGSYR | 180 |
| RDFTVPADWG GRRVELHFDG VNSFFYLWIN GKYVGFSKDS RSTATFDITD YLRKGVNTVA | 240 |
| VEVYRSSDGS FLEAQDMWRL PGIYRSVYLT SSPEVAVRDI AVTPDLDESY RDGTLHIRTE | 300 |
| LRNRSSRTAK GWRVRYSLYQ NTLYSQENSP TGVVVTTDLP GTPRGEASVT ETTLSLRNAE | 360 |
| LWSAERPHLY TLVGELLDPR GRVQQTFSVI TGFRKIEIRD TPAEEDEFGL AGRYFYLNGK | 420 |
| PIKLKGVNRQ EISPETGNTI TREQMIDEIM LMKRGNINHV RLSHYPNTPI FYYLCNLFGL | 480 |
| YLEDEANCES HAYYYGEASL SHVPEFTNAT VQRTMNMVAA HVNEPSILIW SLGNEAGPGK | 540 |
| CFVAAYDAIK AFDKSRPVQY ERNNDIVDIG SNQYPSIAWM REAVQGKYDL KYPFHVSEYA | 600 |
| HSMGNAVGNL IDYWEAIEST NFFMGGAIWD WVDQALWTTD PVTGDRYLAY GGDFGDKPNS | 660 |
| GMFCMNGILL PTHEPKPAYY EVRKVYQDVG ISNEALAEEG TVRIFNKRYF TTLDDLYLEA | 720 |
| VLLEDGREVR TEQVEMPSIA PREAAVVTLP FTRKDTKRDH EYCVTIRLRQ RYETPWAEAG | 780 |
| YVQMAEQLEQ SPAVPSAKIF DGKVQLTLDD SSTISGEGFA VRFDDRKGTI DRLVYGGREV | 840 |
| IASGNGPRLD PFRAPTDNDN WASTAWFANG LHNLRHRVLS RDVHSNADGS LSILYRVESR | 900 |
| APYGTDFSGG VSGRFELKDH PEKPFGPEDC TFASTYIWTV SPDGTITLSA SVESNKPSLA | 960 |
| LARVGFSMRL PRELNRYTYY GRGPWNNYAD RETGAFVGEY SSSVAEQFVP FPKPQSMGNR | 1020 |
| EGVRYGRLTD ETGERGLMIM THGGSMSVLP YSQMQLAMAP HPYQLPVSDG TYVHMDLAVT | 1080 |
| GLGGNSCGQG GPLAPDRVMG GPHTMTLQIR PLPAEGVVAT PTALPVTMRR SATGLLTLSG | 1140 |
| SSPIRYSVDG KPAVRYERPI DLRSGGVVRA WPEGSDWIVT EHRYDRITTI PVTISGVSSE | 1200 |
| ESGAGAVAHL LDGDPSTIWH SMYSVTVAAY PHWVALDCGE ERTIAGITYL PRQDGSTNGD | 1260 |
| IRDYALSVSS DGEKWQEVVR GTMERSKSRQ TLRLSHPVRA RYIRFTALSS QSGADFAAGA | 1320 |
| ELTVLSE | 1327 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 69 | 997 | 5.5e-211 | 0.9747340425531915 |
| CBM32 | 1198 | 1322 | 3.3e-24 | 0.9032258064516129 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK09525 | lacZ | 0.0 | 42 | 1084 | 17 | 999 | beta-galactosidase. |
| PRK10340 | ebgA | 0.0 | 42 | 1089 | 4 | 978 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| COG3250 | LacZ | 2.23e-129 | 73 | 995 | 5 | 804 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| pfam02836 | Glyco_hydro_2_C | 1.08e-75 | 415 | 691 | 8 | 302 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| smart01038 | Bgal_small_N | 1.23e-72 | 815 | 1104 | 1 | 267 | Beta galactosidase small chain. This domain comprises the small chain of dimeric beta-galactosidases EC:3.2.1.23. This domain is also found in single chain beta-galactosidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRO23809.1 | 0.0 | 15 | 1327 | 14 | 1348 |
| QCQ56198.1 | 0.0 | 1 | 1327 | 1 | 1341 |
| QCQ38420.1 | 0.0 | 1 | 1327 | 1 | 1341 |
| AUI47491.1 | 0.0 | 1 | 1327 | 1 | 1341 |
| QTO25654.1 | 0.0 | 1 | 1327 | 1 | 1341 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6S6Z_A | 3.67e-182 | 41 | 1128 | 3 | 998 | Structureof beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_B Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_C Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_D Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_E Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_F Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_G Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_H Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8] |
| 6SD0_A | 3.77e-182 | 41 | 1128 | 4 | 999 | Structureof beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_B Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_C Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_D Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8] |
| 3DEC_A | 3.49e-172 | 41 | 1112 | 6 | 997 | ChainA, Beta-galactosidase [Bacteroides thetaiotaomicron VPI-5482] |
| 3BGA_A | 3.07e-166 | 41 | 1090 | 9 | 980 | Crystalstructure of beta-galactosidase from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482],3BGA_B Crystal structure of beta-galactosidase from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482] |
| 4V41_A | 3.51e-127 | 34 | 1085 | 8 | 996 | E.COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) [Escherichia coli BL21],4V41_B E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) [Escherichia coli BL21],4V41_C E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) [Escherichia coli BL21],4V41_D E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) [Escherichia coli BL21],4V41_E E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) [Escherichia coli BL21],4V41_F E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) [Escherichia coli BL21],4V41_G E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) [Escherichia coli BL21],4V41_H E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) [Escherichia coli BL21],4V41_I E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) [Escherichia coli BL21],4V41_J E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) [Escherichia coli BL21],4V41_K E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) [Escherichia coli BL21],4V41_L E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) [Escherichia coli BL21],4V41_M E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) [Escherichia coli BL21],4V41_N E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) [Escherichia coli BL21],4V41_O E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) [Escherichia coli BL21],4V41_P E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) [Escherichia coli BL21],4V44_A E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH 2-F-LACTOSE [Escherichia coli],4V44_B E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH 2-F-LACTOSE [Escherichia coli],4V44_C E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH 2-F-LACTOSE [Escherichia coli],4V44_D E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH 2-F-LACTOSE [Escherichia coli],4V44_E E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH 2-F-LACTOSE [Escherichia coli],4V44_F E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH 2-F-LACTOSE [Escherichia coli],4V44_G E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH 2-F-LACTOSE [Escherichia coli],4V44_H E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH 2-F-LACTOSE [Escherichia coli],4V44_I E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH 2-F-LACTOSE [Escherichia coli],4V44_J E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH 2-F-LACTOSE [Escherichia coli],4V44_K E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH 2-F-LACTOSE [Escherichia coli],4V44_L E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH 2-F-LACTOSE [Escherichia coli],4V44_M E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH 2-F-LACTOSE [Escherichia coli],4V44_N E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH 2-F-LACTOSE [Escherichia coli],4V44_O E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH 2-F-LACTOSE [Escherichia coli],4V44_P E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH 2-F-LACTOSE [Escherichia coli],4V45_A E. COLI (lacZ) BETA-GALACTOSIDASE-TRAPPED 2-F-GALACTOSYL-ENZYME INTERMEDIATE [Escherichia coli],4V45_B E. COLI (lacZ) BETA-GALACTOSIDASE-TRAPPED 2-F-GALACTOSYL-ENZYME INTERMEDIATE [Escherichia coli],4V45_C E. COLI (lacZ) BETA-GALACTOSIDASE-TRAPPED 2-F-GALACTOSYL-ENZYME INTERMEDIATE [Escherichia coli],4V45_D E. COLI (lacZ) BETA-GALACTOSIDASE-TRAPPED 2-F-GALACTOSYL-ENZYME INTERMEDIATE [Escherichia coli],4V45_E E. COLI (lacZ) BETA-GALACTOSIDASE-TRAPPED 2-F-GALACTOSYL-ENZYME INTERMEDIATE [Escherichia coli],4V45_F E. COLI (lacZ) BETA-GALACTOSIDASE-TRAPPED 2-F-GALACTOSYL-ENZYME INTERMEDIATE [Escherichia coli],4V45_G E. COLI (lacZ) BETA-GALACTOSIDASE-TRAPPED 2-F-GALACTOSYL-ENZYME INTERMEDIATE [Escherichia coli],4V45_H E. COLI (lacZ) BETA-GALACTOSIDASE-TRAPPED 2-F-GALACTOSYL-ENZYME INTERMEDIATE [Escherichia coli],4V45_I E. COLI (lacZ) BETA-GALACTOSIDASE-TRAPPED 2-F-GALACTOSYL-ENZYME INTERMEDIATE [Escherichia coli],4V45_J E. COLI (lacZ) BETA-GALACTOSIDASE-TRAPPED 2-F-GALACTOSYL-ENZYME INTERMEDIATE [Escherichia coli],4V45_K E. COLI (lacZ) BETA-GALACTOSIDASE-TRAPPED 2-F-GALACTOSYL-ENZYME INTERMEDIATE [Escherichia coli],4V45_L E. COLI (lacZ) BETA-GALACTOSIDASE-TRAPPED 2-F-GALACTOSYL-ENZYME INTERMEDIATE [Escherichia coli],4V45_M E. COLI (lacZ) BETA-GALACTOSIDASE-TRAPPED 2-F-GALACTOSYL-ENZYME INTERMEDIATE [Escherichia coli],4V45_N E. COLI (lacZ) BETA-GALACTOSIDASE-TRAPPED 2-F-GALACTOSYL-ENZYME INTERMEDIATE [Escherichia coli],4V45_O E. COLI (lacZ) BETA-GALACTOSIDASE-TRAPPED 2-F-GALACTOSYL-ENZYME INTERMEDIATE [Escherichia coli],4V45_P E. COLI (lacZ) BETA-GALACTOSIDASE-TRAPPED 2-F-GALACTOSYL-ENZYME INTERMEDIATE [Escherichia coli] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q56307 | 2.06e-181 | 41 | 1128 | 4 | 999 | Beta-galactosidase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=lacZ PE=1 SV=2 |
| O52847 | 1.51e-174 | 33 | 1089 | 11 | 1010 | Beta-galactosidase OS=Priestia megaterium (strain DSM 319 / IMG 1521) OX=592022 GN=bgaM PE=3 SV=1 |
| Q9K9C6 | 1.44e-153 | 48 | 1085 | 16 | 986 | Beta-galactosidase OS=Alkalihalobacillus halodurans (strain ATCC BAA-125 / DSM 18197 / FERM 7344 / JCM 9153 / C-125) OX=272558 GN=lacZ PE=3 SV=1 |
| P06864 | 4.57e-149 | 40 | 1088 | 2 | 977 | Evolved beta-galactosidase subunit alpha OS=Escherichia coli (strain K12) OX=83333 GN=ebgA PE=1 SV=4 |
| P81650 | 1.59e-146 | 41 | 1085 | 12 | 1009 | Beta-galactosidase OS=Pseudoalteromonas haloplanktis OX=228 GN=lacZ PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
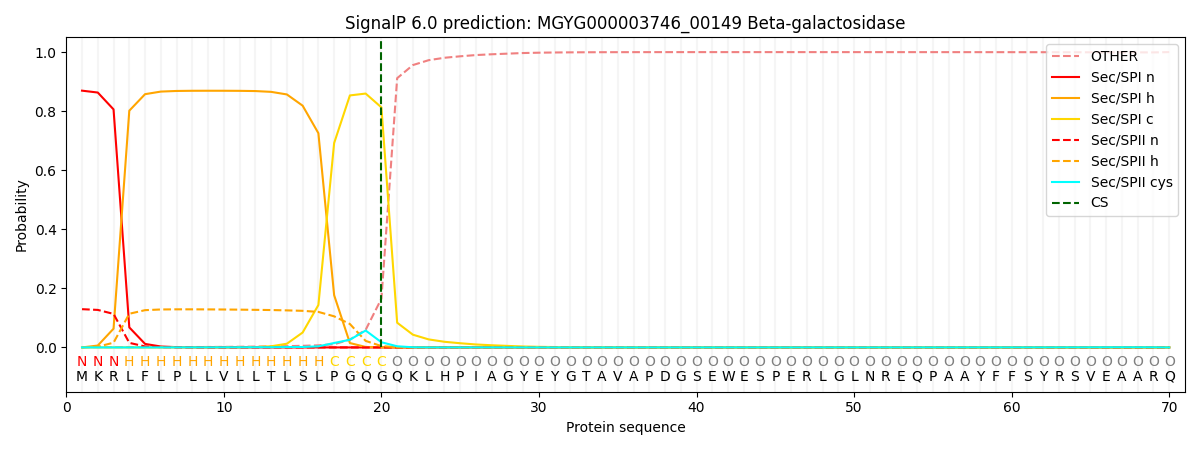
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002828 | 0.862919 | 0.133428 | 0.000319 | 0.000259 | 0.000247 |
