You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003883_00398
You are here: Home > Sequence: MGYG000003883_00398
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Kosakonia cowanii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Kosakonia; Kosakonia cowanii | |||||||||||
| CAZyme ID | MGYG000003883_00398 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 15439; End: 17643 Strand: + | |||||||||||
Full Sequence Download help
| MMRNALLLSL LLSTQAFATA LPVTWSLAGE WRVHDANDPA FDGVSASSAD WRTLRVPANW | 60 |
| YSDGLDHQGA LWYRHSFTLP AQSADTLATL VFDGVDYLAD VTLNGKRLAH HEGYYQRFPV | 120 |
| DVSQALRRHN QLTVRVDSPF EDPKTIWPLH KTMVKGVLNQ HDTRPGGAWS AQGQDANSGG | 180 |
| IWAPVRLHLS RGAAIDEVIL RPQWQQGLTK PMLRAEIRYR AKQAGPVTLR LTAKPDNFYG | 240 |
| RDYRQEFPVT LEAGDSKVLT VTLPMEAARL WWPVGHGKPN LYRVRASLID RQGVMDTAVT | 300 |
| RTGLRKMEEA PDNKGWRIND KRLFVKGSNY IGSPWLGTMT RARYRQDFEL VTRMNANAIR | 360 |
| VHGHVAGRAL YDVADEMGLM IWQDVPLQWG YNNSSEFADN AARQTREMVA QFGNSPAIVA | 420 |
| WGGHNEPPWN SPWMEKRFKD WNKDLNRELT QKVADALAED TSRIVHRYSA VEEHYWQGWY | 480 |
| FGTMRDVLAP AKTGIITEFG AQALPRLSTL KTIIPASEMW PKSTDAKDPG WDRWKYHNFQ | 540 |
| PFQTFGFANV PRGENIQQTI DNTQRYQAEL VAMAAESYRR QRYQPVTALF HFMFVETWPS | 600 |
| INWGVVDYLR QPKAGYYALQ TAYQPLLPSI EPVTANWQTN KANILRLWAI NDSWSACEKC | 660 |
| TLKWRITQND NSLESGDLLL TLDADSGQMV KEVSVTPTAP GEARIAYWIE DQQGKVVGKN | 720 |
| ARVEVVTAGE NAAP | 734 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 26 | 709 | 2.3e-100 | 0.7087765957446809 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 2.84e-44 | 26 | 629 | 14 | 635 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 1.10e-16 | 66 | 427 | 62 | 418 | beta-D-glucuronidase; Provisional |
| PRK10340 | ebgA | 2.05e-11 | 27 | 466 | 44 | 473 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| pfam00703 | Glyco_hydro_2 | 3.08e-09 | 213 | 305 | 20 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| pfam02837 | Glyco_hydro_2_N | 5.18e-09 | 26 | 136 | 3 | 133 | Glycosyl hydrolases family 2, sugar binding domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities and has a jelly-roll fold. The domain binds the sugar moiety during the sugar-hydrolysis reaction. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AST70760.1 | 0.0 | 1 | 734 | 1 | 734 |
| QAR44936.1 | 0.0 | 1 | 734 | 1 | 734 |
| APZ04632.1 | 0.0 | 1 | 728 | 1 | 728 |
| AZI86236.1 | 0.0 | 1 | 728 | 1 | 728 |
| QNQ21798.1 | 0.0 | 1 | 734 | 1 | 734 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5N6U_A | 1.51e-32 | 90 | 632 | 102 | 662 | Crystalstructure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12],5N6U_B Crystal structure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12],5N6U_C Crystal structure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12],5N6U_D Crystal structure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12] |
| 6BYE_A | 8.62e-32 | 26 | 625 | 7 | 672 | Crystalstructure of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri in complex with mannose [Xanthomonas citri pv. citri str. 306],6BYE_B Crystal structure of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri in complex with mannose [Xanthomonas citri pv. citri str. 306] |
| 6BYC_A | 8.63e-32 | 26 | 625 | 7 | 672 | Crystalstructure of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306] |
| 6BYI_A | 2.00e-31 | 26 | 625 | 5 | 670 | Crystalstructure of the acid-base mutant (E477A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306],6BYI_B Crystal structure of the acid-base mutant (E477A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306] |
| 6BYG_A | 4.67e-31 | 26 | 625 | 7 | 672 | Crystalstructure of the nucleophile mutant (E575A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306],6BYG_B Crystal structure of the nucleophile mutant (E575A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q4WAH4 | 6.78e-34 | 73 | 648 | 68 | 683 | Beta-mannosidase B OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=mndB PE=3 SV=1 |
| B0YBU9 | 1.20e-33 | 73 | 648 | 68 | 683 | Beta-mannosidase B OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=mndB PE=3 SV=1 |
| A1CGA8 | 1.59e-33 | 73 | 648 | 68 | 683 | Beta-mannosidase B OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=mndB PE=3 SV=1 |
| A1D911 | 8.73e-33 | 73 | 648 | 68 | 683 | Beta-mannosidase B OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=mndB PE=3 SV=1 |
| Q0CCA0 | 1.15e-32 | 26 | 652 | 8 | 685 | Beta-mannosidase B OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=mndB PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
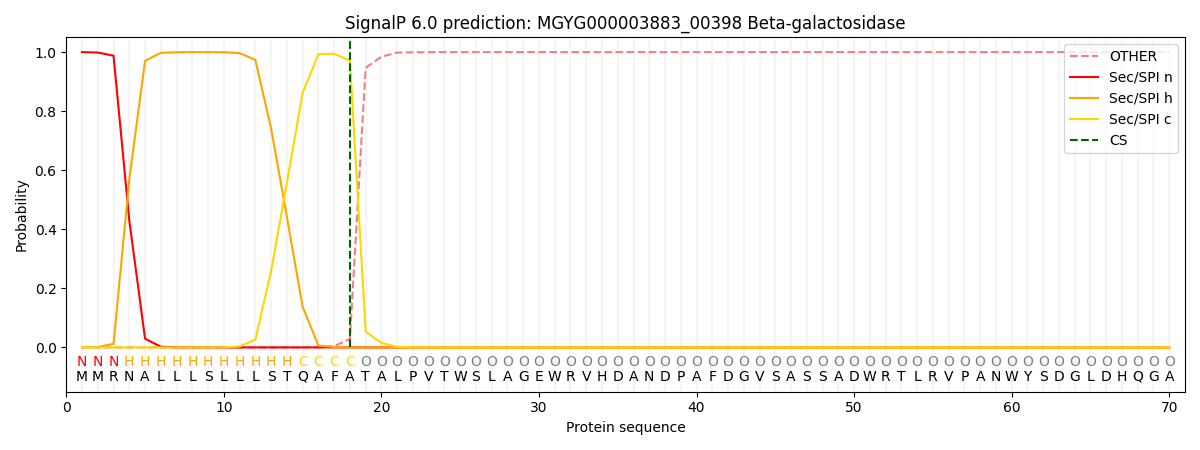
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001033 | 0.997213 | 0.000510 | 0.000439 | 0.000379 | 0.000372 |
