You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003922_00499
You are here: Home > Sequence: MGYG000003922_00499
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
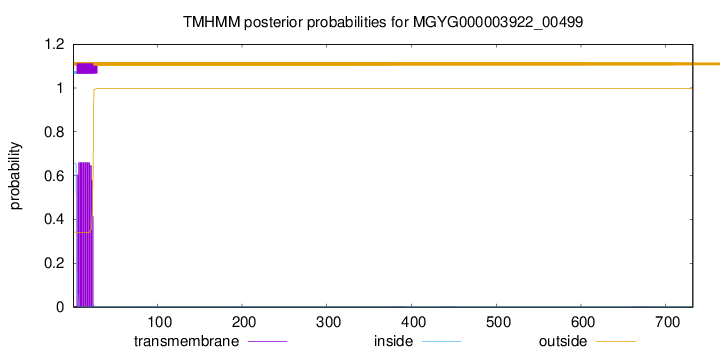
TMHMM annotations
Basic Information help
| Species | Bacteroides sp014385165 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides sp014385165 | |||||||||||
| CAZyme ID | MGYG000003922_00499 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 131955; End: 134153 Strand: - | |||||||||||
Full Sequence Download help
| MKQNWILIAM LLYVIPCLAW EPVGDKIKTR WAKEVTPDNV WKEYPRPQLR RADWQNLNGL | 60 |
| WQYTIIGKNE KAPKTYSGEI LVPFCVESSL SGVGKTVRPE DKIWYKTTFE IPTDWKGENI | 120 |
| LLHFGAVDWE TTVWVNGKQA GTHKGGFDAF SFDITKCLKG SGKQELVVSV WDPTDFGSQA | 180 |
| RGKQQLNQQG IWYTPVSGIW QTVWLEPVNK SHIASVTPVA DIDHRSVTLK TAVSNTSKSD | 240 |
| KLQISVKDGG KTILTKEMAY QPELTLNIPE PKLWTPATPH LYQLEMTLKR GNKTLDKVDS | 300 |
| YFAMRKVSKA RDEKGYMRIC LNNEPIFQYG TLDQGWWPDG LHTPPTAEAM RWDMETLKEM | 360 |
| GFNSLRKHIK MEPALYYYYA DSLGIMIWQD MSSGMSSQNK AQEHVKPGAA KDWESRPAES | 420 |
| AQEWESELKR MIDQLGFFPC ITTWVVFNEG WGQYDTPRIV NWVMDYDKTR LINGVSGWAD | 480 |
| RGVGHFYDYH NYPAASMPLP KDCGERVSVL GEFGGLGLPL KEHLWNASMR NWGYKTINES | 540 |
| NTLINDYTRL MYDLRTLAAT GLSAAIYTQT TDVEGEVNGL ITYDREVIKI PAPMLHAIHS | 600 |
| ELYNAKTSAV NNLIADARYG KTTHLFQYGS APAQKVSFPV NIKQKTNVTS TEEFTLNKIP | 660 |
| SNLMIWLNMS GNTTVWLNGH RVLDQNVRQT RQYNQFNLSD YIPYLKVGKN TLKVECDAQN | 720 |
| DKLFDYGLQA ID | 732 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 43 | 576 | 1.6e-105 | 0.5930851063829787 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 5.91e-37 | 55 | 473 | 13 | 430 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 1.18e-29 | 56 | 449 | 14 | 417 | beta-D-glucuronidase; Provisional |
| PRK10340 | ebgA | 1.03e-18 | 52 | 326 | 69 | 334 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| pfam02837 | Glyco_hydro_2_N | 1.35e-13 | 55 | 205 | 2 | 165 | Glycosyl hydrolases family 2, sugar binding domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities and has a jelly-roll fold. The domain binds the sugar moiety during the sugar-hydrolysis reaction. |
| PRK09525 | lacZ | 3.99e-13 | 105 | 366 | 124 | 389 | beta-galactosidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNL39997.1 | 4.88e-304 | 19 | 730 | 21 | 733 |
| QUT79147.1 | 3.23e-302 | 19 | 730 | 21 | 733 |
| QDM10564.1 | 3.23e-302 | 19 | 730 | 21 | 733 |
| ALJ47794.1 | 4.58e-302 | 19 | 730 | 21 | 733 |
| QRQ54685.1 | 4.58e-302 | 19 | 730 | 21 | 733 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7SF2_A | 2.68e-207 | 20 | 605 | 5 | 585 | ChainA, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_B Chain B, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_C Chain C, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_D Chain D, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_E Chain E, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_F Chain F, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838] |
| 4YPJ_A | 5.18e-25 | 47 | 470 | 32 | 437 | ChainA, Beta galactosidase [Niallia circulans],4YPJ_B Chain B, Beta galactosidase [Niallia circulans] |
| 6S6Z_A | 5.20e-25 | 105 | 499 | 113 | 491 | Structureof beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_B Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_C Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_D Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_E Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_F Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_G Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_H Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8] |
| 6SD0_A | 5.21e-25 | 105 | 499 | 114 | 492 | Structureof beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_B Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_C Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_D Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8] |
| 7CWD_A | 2.78e-24 | 42 | 470 | 21 | 431 | ChainA, beta-glalactosidase [Niallia circulans],7CWI_A Chain A, beta-galactosidase [Niallia circulans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KM09 | 6.92e-29 | 104 | 477 | 109 | 459 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22050 PE=2 SV=2 |
| Q56307 | 2.85e-24 | 105 | 499 | 114 | 492 | Beta-galactosidase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=lacZ PE=1 SV=2 |
| P77989 | 6.69e-22 | 104 | 475 | 59 | 415 | Beta-galactosidase OS=Thermoanaerobacter pseudethanolicus (strain ATCC 33223 / 39E) OX=340099 GN=lacZ PE=3 SV=2 |
| T2KPJ7 | 5.30e-21 | 104 | 391 | 106 | 389 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_21970 PE=2 SV=1 |
| Q6D736 | 4.65e-15 | 37 | 480 | 41 | 503 | Beta-galactosidase OS=Pectobacterium atrosepticum (strain SCRI 1043 / ATCC BAA-672) OX=218491 GN=lacZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
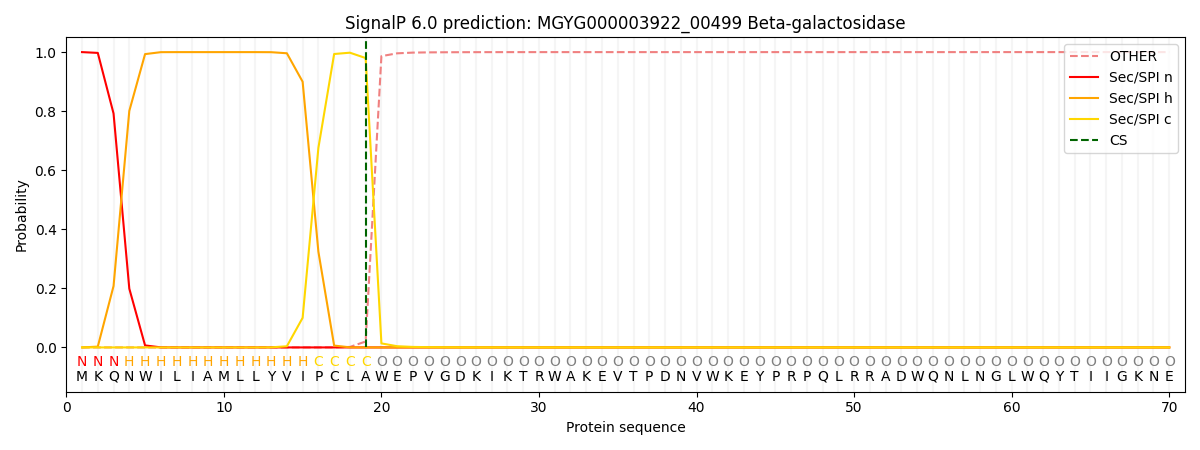
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000208 | 0.999222 | 0.000147 | 0.000140 | 0.000129 | 0.000128 |