You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001442_00220
You are here: Home > Sequence: MGYG000001442_00220
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Streptococcus sp000411475 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Streptococcaceae; Streptococcus; Streptococcus sp000411475 | |||||||||||
| CAZyme ID | MGYG000001442_00220 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 220135; End: 228354 Strand: + | |||||||||||
Full Sequence Download help
| MGKHFFERRC HYSIRKFAIG AASVMIGASI FGAGVVQAAE TEGPAETEGT VTQVQPMDKL | 60 |
| PADIAAAIEK AETASPSEAT ESQPTEPATQ PANTGTVTPE AKPAETPVPK EEATPKPAET | 120 |
| PKEEAAPVAK PAETPVAKDV VETPDVNHLE KATATVSNHE ANTPFTAEKA IDGNPDTRWA | 180 |
| TDRDVVKPTI EFKLEKTTLI KHVEIDWDRR VRGEQNDPNI KSWNLYYAGQ DNVNGSGPGE | 240 |
| WKLAHQRTGT PVLDEKVDLK EAVQAKYLKL EITDYQAGTM QWKNVGIQEI RAYSNIPDTS | 300 |
| KPTDIRQVTE LAVAEDGKSL VLPKLPGQVS LIGSNKQGVV DLNNKIYTPL TEQHVKVMVQ | 360 |
| QTNDNHTFTK EFEVVIKGLH ADEGVGTKPA VAPAVQQWYG TEGKTSITSE TVISVGNSGF | 420 |
| DKEAKFYQTD LENRGLEVAT GSQEAKNRIE FKKVEDKGYG KEGYGITIKD GVITVEAATN | 480 |
| AGAFYATRTL LQLGENNLQN GEIRDFPSFS HRGFMLDTGR KFIPYDTLVD IMLNMAYYKM | 540 |
| NDLQLHLNDN YIFLKKHVEG KHLSQQGELD YVLKNAKTGF RVETDVVGDN GEKLTSKEHY | 600 |
| TKDELQQIIS LAKDLHINLV PEIDTPGHAL SFVKVRPDLM YKGPLSANKH NVERVAMLDL | 660 |
| DNKYEETLAF VKSVYDKLLV GENAPLRGVS TVHIGTDEYY GSPENYRRYV HDMIQYIKDK | 720 |
| GLTPRIWGSL TAKPGKTPVD WNGVEVDIWS LGWQNPQAAI AKGAKIINIL DVPTYSVPNG | 780 |
| SNSQGPYSDY ANYELQYNSW APNDFTARRG PRLEASNPNI IGGGHAVWND NIDLHETGLT | 840 |
| SFDIFKRFFK SMQSTAERTW GSDRAAKTYA DRIQPTSVYA PRSNPEKTIE DSDLFTIKPE | 900 |
| TIKEYLAKNV KKTEAGLNFE KDSSIEGLVG DVGPSHVLKL DVTVTGDGEQ VFSTSGDNQL | 960 |
| YLADKDGYLA YKFEQFHIQF DKKLEKNKRY QISVVTKPQK TEVYVDGEKV ERIANPAHPR | 1020 |
| LAHNSLVLPL ETIGGFQGIL HSAELSNEAF VNPRLIPTDH FTVSATSQET PGTETEGPVE | 1080 |
| KAFDNDPNTF WHSKWTGHQA PFTVAMNLKA PEKVNGLTYL PRPGGGNGVV TSYEIYAQKD | 1140 |
| GQMVKVASGT WENNTKEKTV NFAAIETNKV EFKVLSGFAG FGSAAEIQLL KPLSDSESEE | 1200 |
| PVAPEKPVTP EKPVTPEKPK VEEVSDGTTE LADSFVATKP ASDDAIAAAT QSQDYLKKEY | 1260 |
| KVFPTPQKVT YGEGVTKLQK QVNLVMGDHL DIYTRNRLKS VLQDHQISYT SSQAAVAGAT | 1320 |
| NIYLGVHGQH SQAEKEISGI SQGLFDKIDA YALSIKNNTI SIVGKDTDAV FYGLTTLKHM | 1380 |
| LNESEAPVLR NVTVEDYAEI KNRGFIEGYY GNPWTNADRA ELMRYGGDLK MTQYFFAPKD | 1440 |
| DPYHNKKWRE LYPEEKLAEI RELARVGNQN KTRYVWTIHP FMNNRIRFGN DADYQKDLET | 1500 |
| IKAKFTQLMD AGVREFGILA DDAPSPVGGY NSYNRLMKDM TDWLTEKQAT YVGLRKEMIF | 1560 |
| VPGQYWGNGR EDELKSLNEN LPSSTSMTLT GGKIWGEVSE NFLSNLKNNL TAGGKTYRPV | 1620 |
| SLWINWPCTD NSKQHLILGG GEKFLHPNVD PSLLSGIMLN PMQQSEPSKI ALFSAAQYAW | 1680 |
| KQWKSEDEAK KVNDIAFNFV ETGKFTDSET SVAFRELGKH MINQNMDGRV VKLEESVELA | 1740 |
| PKLDAFMAKL KAGQDVSAER AELRAEFAKL KAAAQLYKAS GDEKMRSQIH YWLDNTIDQM | 1800 |
| DALSAFLDGS EAIENNDSAR LWDSYYKGLK LYEQSQTYTF HYVDHDERAE LGVQHIRPFL | 1860 |
| LGLREVLATE VQKALHPDQV ISTFITNRTG VEGGLAEVTD GDLGTHALIK SPNSIQTGDY | 1920 |
| IGLKFNKAVP IQNLTFAMGT QANPHDTFNN AKVEYLNEND EWVTLSEPSY TGNEPLLKFE | 1980 |
| NLNINAKAVR MIATSDRDNT WFAVREIAVN RPVEVSRPKQ AVTVTISPNL MYKYNTTVAQ | 2040 |
| ITDGRDNTEA MLANADRTDS TPVDGWVQLD LGGVKPVTKV RLVQGSGDKL AEGVLEYSTD | 2100 |
| GSSWQELDRL TGEQTKEIET PISARYIRVR NTKNINLWWR IADFSVETRT GNSEMTDTNV | 2160 |
| ESLKSTPVYD SLGRYDMQIP SGTKLPAHSY LGMKLDRLHQ AESIQAIGIG NPSLDLEYSP | 2220 |
| NAQEWYPASQ VTDKSLVRYA RLVNKTDQEQ AVTATSLLVK TKEVQPTKLD STSMGIDAHY | 2280 |
| GANDVRKIKN LDQLFDGVYN NFVEFSDYAH KDGHITLKLG SEREIKKIRA YIQDGTKNYL | 2340 |
| RDGKIQVSQD GKTWTDVVTV GDGIANDMHD DSLTDGWTHD SKMPGNRYIE GELASPVKAN | 2400 |
| YLRVLFTANY DARFVGFTEL VINDGEFVKP TNDPTVQGNS GESRGNLYTN LVDGKVLNSY | 2460 |
| KAEKDQGELV YHLSEPTDAN HIRLVSSLPQ GVAARVLART LKSDRDGAWT DLGAITSSFQ | 2520 |
| TFAVREKAPL LDVKLIWEGG KPEFYEMTTF HQELSEEPEQ PTPDPEPTPT PEPTPETPTS | 2580 |
| SKGEEQPPVV EIPEYTEPIG TAGDQAAPVV EIPKFTGGVN AVEAAKNEVP EFKGGVNWVE | 2640 |
| ADKNELPEFK GGVNWVEAAK NEVPEYKEPV ATPAEPLADH KPTPSATNQK PEQEGPNAPA | 2700 |
| TGAKAQLPAT GEQDSVGLAF IASLGLVLSA TFIKKGKED | 2739 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH84 | 1403 | 1702 | 1.8e-97 | 0.9559322033898305 |
| GH20 | 506 | 861 | 3.9e-59 | 0.9643916913946587 |
| CBM32 | 155 | 289 | 4.9e-17 | 0.9193548387096774 |
| CBM32 | 1065 | 1187 | 5.2e-16 | 0.9516129032258065 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07555 | NAGidase | 2.63e-102 | 1403 | 1680 | 1 | 263 | beta-N-acetylglucosaminidase. This family has previously been described as a hyaluronidase. However, more recently it has been shown that this family has beta-N-acetylglucosaminidase activity. |
| cd06564 | GH20_DspB_LnbB-like | 4.52e-102 | 511 | 861 | 2 | 325 | Glycosyl hydrolase family 20 (GH20) catalytic domain of dispersin B (DspB), lacto-N-biosidase (LnbB) and related proteins. Dispersin B is a soluble beta-N-acetylglucosamidase found in bacteria that hydrolyzes the beta-1,6-linkages of PGA (poly-beta-(1,6)-N-acetylglucosamine), a major component of the extracellular polysaccharide matrix. Lacto-N-biosidase hydrolyzes lacto-N-biose (LNB) type I oligosaccharides at the nonreducing terminus to produce lacto-N-biose as part of the GNB/LNB (galacto-N-biose/lacto-N-biose I) degradation pathway. The lacto-N-biosidase from Bifidobacterium bifidum has this GH20 domain, a carbohydrate binding module 32, and a bacterial immunoglobulin-like domain 2, as well as a YSIRK signal peptide and a G5 membrane anchor at the N and C termini, respectively. The GH20 hexosaminidases are thought to act via a catalytic mechanism in which the catalytic nucleophile is not provided by solvent or the enzyme, but by the substrate itself. |
| cd02742 | GH20_hexosaminidase | 3.65e-35 | 511 | 861 | 1 | 303 | Beta-N-acetylhexosaminidases of glycosyl hydrolase family 20 (GH20) catalyze the removal of beta-1,4-linked N-acetyl-D-hexosamine residues from the non-reducing ends of N-acetyl-beta-D-hexosaminides including N-acetylglucosides and N-acetylgalactosides. These enzymes are broadly distributed in microorganisms, plants and animals, and play roles in various key physiological and pathological processes. These processes include cell structural integrity, energy storage, cellular signaling, fertilization, pathogen defense, viral penetration, the development of carcinomas, inflammatory events and lysosomal storage disorders. The GH20 enzymes include the eukaryotic beta-N-acetylhexosaminidases A and B, the bacterial chitobiases, dispersin B, and lacto-N-biosidase. The GH20 hexosaminidases are thought to act via a catalytic mechanism in which the catalytic nucleophile is not provided by the solvent or the enzyme, but by the substrate itself. |
| pfam00728 | Glyco_hydro_20 | 6.22e-31 | 509 | 860 | 1 | 343 | Glycosyl hydrolase family 20, catalytic domain. This domain has a TIM barrel fold. |
| COG3525 | Chb | 5.85e-30 | 336 | 761 | 77 | 517 | N-acetyl-beta-hexosaminidase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QLF55290.1 | 0.0 | 1 | 2739 | 1 | 2822 |
| CBJ22777.1 | 0.0 | 1 | 2739 | 1 | 2770 |
| AMD97395.1 | 0.0 | 1 | 2628 | 1 | 2589 |
| AMH89096.1 | 0.0 | 1 | 2739 | 1 | 2766 |
| QUB38718.1 | 0.0 | 1 | 2628 | 1 | 2592 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6PV4_A | 2.69e-188 | 1260 | 1871 | 32 | 648 | Structureof CpGH84A [Clostridium perfringens ATCC 13124],6PV4_B Structure of CpGH84A [Clostridium perfringens ATCC 13124],6PV4_C Structure of CpGH84A [Clostridium perfringens ATCC 13124],6PV4_D Structure of CpGH84A [Clostridium perfringens ATCC 13124] |
| 6PWI_A | 1.23e-107 | 1248 | 1867 | 22 | 624 | Structureof CpGH84D [Clostridium perfringens ATCC 13124],6PWI_B Structure of CpGH84D [Clostridium perfringens ATCC 13124] |
| 6JQF_A | 1.56e-104 | 313 | 1038 | 12 | 724 | Crystallizationanalysis of a beta-N-acetylhexosaminidase (Am2136) from Akkermansia muciniphila [Akkermansia muciniphila ATCC BAA-835] |
| 6PV5_A | 1.77e-61 | 1241 | 1831 | 22 | 596 | Structureof CpGH84B [Clostridium perfringens ATCC 13124] |
| 2V5D_A | 1.56e-53 | 1264 | 1999 | 18 | 722 | Structureof a Family 84 Glycoside Hydrolase and a Family 32 Carbohydrate-Binding Module in Tandem from Clostridium perfringens. [Clostridium perfringens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P26831 | 2.29e-297 | 1260 | 2553 | 39 | 1354 | Hyaluronoglucosaminidase OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagH PE=1 SV=2 |
| B2UPR7 | 9.53e-110 | 313 | 1038 | 34 | 746 | Beta-hexosaminidase Amuc_2136 OS=Akkermansia muciniphila (strain ATCC BAA-835 / DSM 22959 / JCM 33894 / BCRC 81048 / CCUG 64013 / CIP 107961 / Muc) OX=349741 GN=Amuc_2136 PE=1 SV=1 |
| Q8XL08 | 8.72e-52 | 1264 | 2030 | 48 | 781 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagJ PE=1 SV=1 |
| Q0TR53 | 1.15e-51 | 1264 | 1999 | 48 | 752 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain ATCC 13124 / DSM 756 / JCM 1290 / NCIMB 6125 / NCTC 8237 / Type A) OX=195103 GN=nagJ PE=1 SV=1 |
| Q89ZI2 | 3.93e-50 | 1264 | 1784 | 28 | 510 | O-GlcNAcase BT_4395 OS=Bacteroides thetaiotaomicron (strain ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50) OX=226186 GN=BT_4395 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
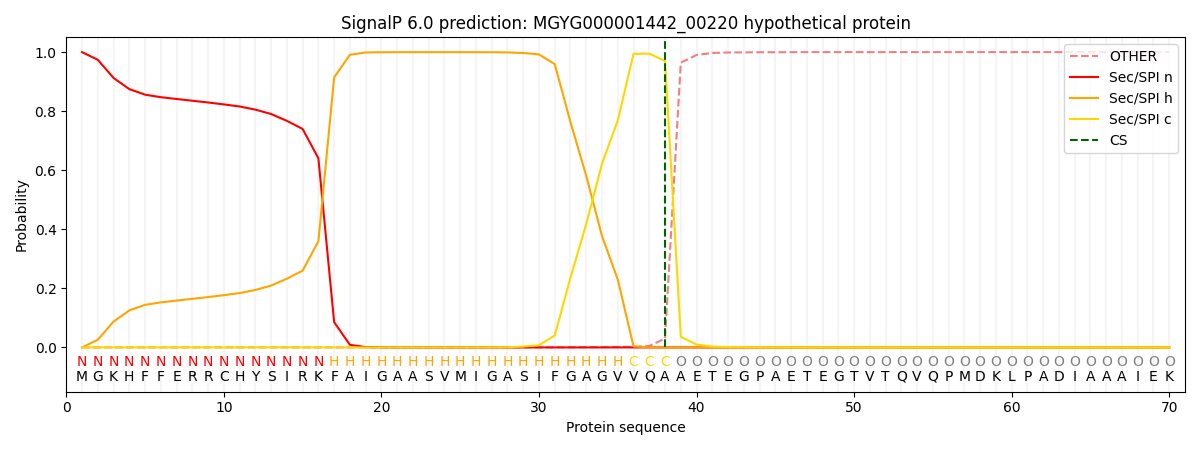
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000433 | 0.998703 | 0.000217 | 0.000240 | 0.000200 | 0.000158 |

