You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000002_01677
You are here: Home > Sequence: MGYG000000002_01677
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
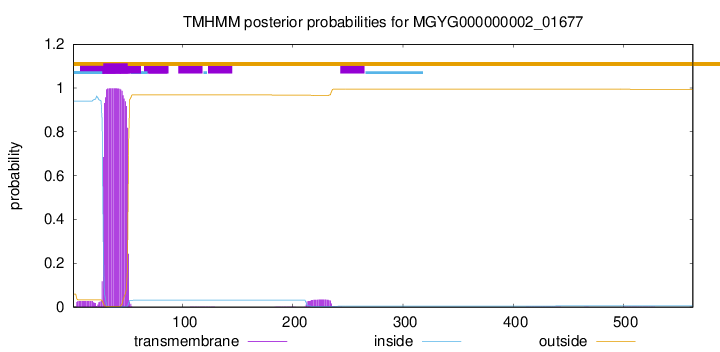
TMHMM annotations
Basic Information help
| Species | Blautia_A faecis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Blautia_A; Blautia_A faecis | |||||||||||
| CAZyme ID | MGYG000000002_01677 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 104363; End: 106054 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd16891 | CwlT-like | 1.07e-55 | 241 | 388 | 1 | 151 | CwlT-like N-terminal lysozyme domain and similar domains. CwlT is a bifunctional cell wall hydrolase containing an N-terminal lysozyme domain and a C-terminal NlpC/P60 endopeptidase domain (gamma-d-D-glutamyl-L-diamino acid endopeptidase), and has been implicated in the spread of transposons. Proteins similar to this family include the soluble and insoluble membrane-bound LTs in bacteria, the LTs in bacteriophage lambda, as well as the eukaryotic "goose-type" lysozymes (goose egg-white lysozyme; GEWL). |
| pfam13702 | Lysozyme_like | 2.23e-51 | 235 | 387 | 1 | 165 | Lysozyme-like. |
| cd02549 | Peptidase_C39A | 1.31e-21 | 425 | 544 | 6 | 126 | A sub-family of peptidase family C39. Peptidase family C39 mostly contains bacteriocin-processing endopeptidases from bacteria. The cysteine peptidases in family C39 cleave the "double-glycine" leader peptides from the precursors of various bacteriocins (mostly non-lantibiotic). The cleavage is mediated by the transporter as part of the secretion process. Bacteriocins are antibiotic proteins secreted by some species of bacteria that inhibit the growth of other bacterial species. The bacteriocin is synthesized as a precursor with an N-terminal leader peptide, and processing involves removal of the leader peptide by cleavage at a Gly-Gly bond, followed by translocation of the mature bacteriocin across the cytoplasmic membrane. Most endopeptidases of family C39 are N-terminal domains in larger proteins (ABC transporters) that serve both functions. The proposed protease active site is conserved in this sub-family of proteins with a single peptidase domain, which are lacking the nucleotide-binding transporter signature or have different domain architectures. |
| pfam13529 | Peptidase_C39_2 | 3.05e-16 | 402 | 531 | 1 | 139 | Peptidase_C39 like family. |
| pfam12385 | Peptidase_C70 | 2.36e-11 | 413 | 532 | 6 | 126 | Papain-like cysteine protease AvrRpt2. This is a family of cysteine proteases, found in actinobacteria, protobacteria and firmicutes. Papain-like cysteine proteases play a crucial role in plant-pathogen/pest interactions. On entering the host they act on non-self substrates, thereby manipulating the host to evade proteolysis. AvrRpt2 from Pseudomonas syringae pv. tomato DC3000 triggers resistance to P. syringae-2-dependent defense responses, including hypersensitive cell death, by cleaving the Arabidopsis RIN4 protein which is monitored by the cognate resistance protein RPS2. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRT48860.1 | 0.0 | 1 | 563 | 1 | 563 |
| QHB23689.1 | 0.0 | 5 | 563 | 38 | 607 |
| ASM68260.1 | 0.0 | 5 | 563 | 38 | 607 |
| QEI31189.1 | 0.0 | 5 | 563 | 38 | 607 |
| CBL21023.1 | 3.33e-310 | 5 | 563 | 38 | 607 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4FDY_A | 8.06e-12 | 232 | 424 | 20 | 216 | ChainA, Similar to lipoprotein, NLP/P60 family [Staphylococcus aureus subsp. aureus Mu50],4FDY_B Chain B, Similar to lipoprotein, NLP/P60 family [Staphylococcus aureus subsp. aureus Mu50] |
| 4HPE_A | 1.91e-10 | 210 | 394 | 2 | 184 | ChainA, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_B Chain B, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_C Chain C, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_D Chain D, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_E Chain E, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_F Chain F, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P96645 | 2.47e-26 | 221 | 386 | 40 | 191 | Probable endopeptidase YddH OS=Bacillus subtilis (strain 168) OX=224308 GN=yddH PE=3 SV=1 |
| O34636 | 6.71e-16 | 234 | 383 | 48 | 200 | Uncharacterized membrane protein YocA OS=Bacillus subtilis (strain 168) OX=224308 GN=yocA PE=4 SV=1 |
| Q99ZN9 | 4.90e-10 | 236 | 365 | 28 | 148 | Pneumococcal vaccine antigen A homolog OS=Streptococcus pyogenes serotype M1 OX=301447 GN=pvaA PE=3 SV=1 |
| Q5XC63 | 1.65e-09 | 236 | 365 | 28 | 148 | Pneumococcal vaccine antigen A homolog OS=Streptococcus pyogenes serotype M6 (strain ATCC BAA-946 / MGAS10394) OX=286636 GN=pvaA PE=3 SV=1 |
| Q8P120 | 1.65e-09 | 236 | 365 | 28 | 148 | Pneumococcal vaccine antigen A homolog OS=Streptococcus pyogenes serotype M18 (strain MGAS8232) OX=186103 GN=pvaA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
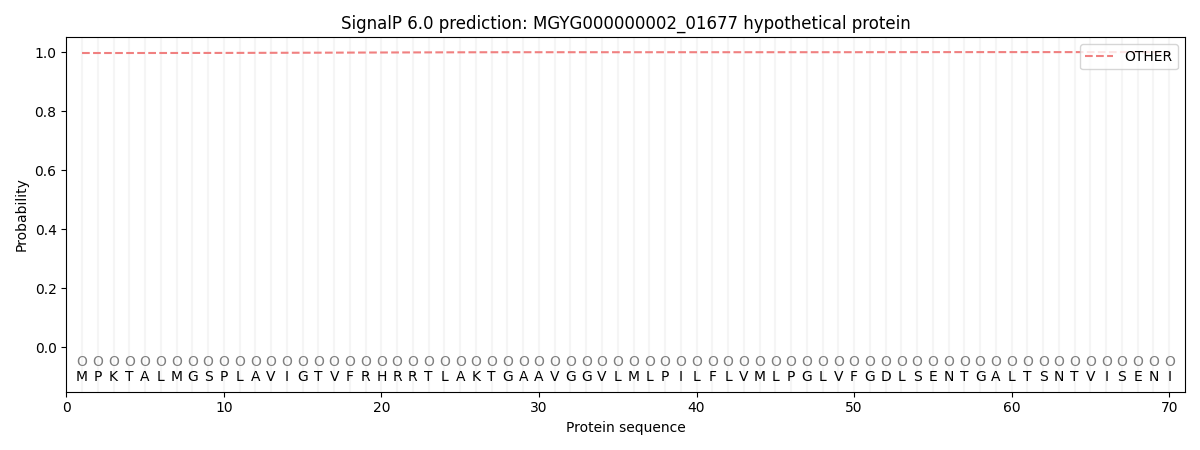
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.997643 | 0.002248 | 0.000015 | 0.000011 | 0.000006 | 0.000085 |