You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000010_01210
You are here: Home > Sequence: MGYG000000010_01210
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
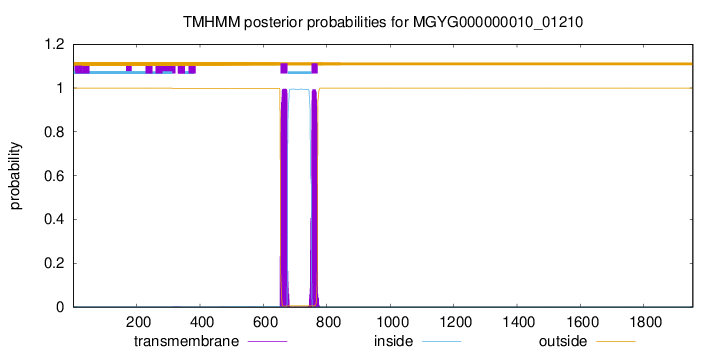
TMHMM annotations
Basic Information help
| Species | Staphylococcus_A lentus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Staphylococcales; Staphylococcaceae; Staphylococcus_A; Staphylococcus_A lentus | |||||||||||
| CAZyme ID | MGYG000000010_01210 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | Chromosome partition protein Smc | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 67197; End: 73064 Strand: + | |||||||||||
Full Sequence Download help
| MANERMEGLT LEMKLDSVGV SEGLKGLKRE LGVVNSEMKA NLSAFDKSDR SMQKYQTRLD | 60 |
| GLNTKLTVQK KMYSQAKQEL KDLNTSYKNA GTEIKKVETQ LSKLTDEHKK NDQALKKSND | 120 |
| EIKKSNDGLK KARMQQKVVN DEKTKAKQKL DQLRQTEMQL RNSGKATTDQ IKKASNATKQ | 180 |
| QRQVHQNLVE QYKKERATVK QLSDTHKRLK GENDKVKQSY KQSNAELKNT QNQHKKLNKE | 240 |
| IQDYTKNQAE ATKKINNEKA SLNNLEKAIE KTSNEIKLFN KEQALANSHF TKTADKFDKL | 300 |
| SDSYSRFGMG LQSVGRNMSM YVTTPIVGAL GYASKLGVEF DDSMRKVQAT SGASGKQLDQ | 360 |
| LKAKAREMGA TTKFSATDSA EALNYMALAG WKTNDMVKGL PGIMNLAAAS GEDLAQVSDI | 420 |
| VTDGLTAFGL KAKDSGRFAD VLAAASANAN TNVLGMGEAF KYAAPVAGAL GYTIEDTSIA | 480 |
| IGLMSNAGVK GQKAGTALRT MFTNLAKPTK AMKNQMEELG ITITDSNGEM LPMRDVLDQL | 540 |
| REKFSGLSKD QQASAAATIF GKEAMSGALA VINASESDYT KLSKAIDKSE GSATAMSKTM | 600 |
| ESGLGGSLRE LRSAAEELGL SIFESVQPAL KGMVSGLKNT VDYLNGLPKG AKLAAVGFGV | 660 |
| ITAAIGPLLL GTGLLIRGIG SAMKGYAGLN RVMAKNSVEA GINAAAHSAA GTSMVKTTAK | 720 |
| NNKGLSRFTS LIGLSGKSTG KLSTVLRVGS KGLGVFGLAI TAVAGTLTYA YKNIDWFRSG | 780 |
| VQSLGTFLKE VGSSIDWSWI SDSAKWFKKA GSATKDFGTK LLELNPYMQL AKLYFGKLDD | 840 |
| AVSKSTDTVD IYGEGVSKGT KKALSKYTDL SEKAKVKLEE IRISHDVIGK EQYKTVSDLY | 900 |
| SNINKEVENQ LEQRHDKEIG KLTKLFKESD ALSGKREQEI LQKTKDAHQK ELDETQKVND | 960 |
| QIQEIYKTAK DEKRALTEKE NEKIASLQSQ LDIKVTESLS NGEKEQKVIL ERMKNNQSAL | 1020 |
| SMEAASNVIK ESAKSRDKSI GDAKKKRDDV IKWAIEQRDE AKTLSKDEAD KIIKDANDKY | 1080 |
| KESKDKAKLQ HKKVVSEAQK QNKGVKENID SQNGHVLSSW EKTKNKVSKH AGKMAGSAIG | 1140 |
| SFLRTSKGAT KWLGKATGTA VSKFKSISKG AGDWLGRAAG TAAGKFTSIS SSAAKWLGKT | 1200 |
| AGNALSKFTN VSSSAGKWLG KTAGTATSKF TTVWNSAKDR FNRTRDKAES STKSVSKSAG | 1260 |
| KWYGSAAGTA KSRFGEIWGS AKSNFTNVAE HGWNKAKSTY NGFKTWLGRT LKYIRDVGSD | 1320 |
| MGRAASDLGI KVANNAIEGL NKMIAGVNKI SAAITGDNLI KTIDTIGSAG TLGRRLSTGT | 1380 |
| VGGDVSTDSD GALKQATMAI VNDKGPGNGT GGFTQEIIER KDGSMYMPKG KDVPVMLEPG | 1440 |
| SKVHSGSVTQ QLAKLGLLPK FSTGTPRKKR LDEQLVGAFG KLTGKTVGHG AEALDGIGTL | 1500 |
| TDKAKGEAGK QAGKASAFVS KSVKDVWDFV DKPKELIKRV MDDFGVNFKS IPAHTGELVR | 1560 |
| GAYKKLKSSL ADKVTSMFDE FGSSGGGYNP FADWPITPGR GWKSGGHAGI DYAVPAGTKI | 1620 |
| PSPLDGEVIQ RWFSPYGGGN ETQVYDGSKY THIFMHQSKQ IAKKGQRINQ GDIIGLVGNT | 1680 |
| GNSFGAHLHW QVNKGKGFRN NHPDSINPLD WVKEAMAAGG AGGKGAAYAR KIIKQAQNIL | 1740 |
| GGRYKSPAIT EHMMKLAKRE SNFDPKAVNN WDSNAQRGTP SKGMFQMIEP TFRANAKSGY | 1800 |
| NKFGNTLHEA ISSLQYIVRT YGWGGFPRAA SAAYANGGIS STHKIAQISE GNKAEAIVPL | 1860 |
| TKRTRAIQLT EQIMDYLGMD TGKSNVTVNN DTSVMEKLLK QLVSLNDKSN RQTDVIIKLM | 1920 |
| KQIPEGTDIS KLEPMISTLQ GSRLNNANYT MGGAR | 1955 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| TIGR01760 | tape_meas_TP901 | 5.68e-71 | 323 | 665 | 1 | 345 | phage tail tape measure protein, TP901 family, core region. This model represents a reasonably well conserved core region of a family of phage tail proteins. The member from phage TP901-1 was characterized as a tail length tape measure protein in that a shortened form of the protein leads to phage with proportionately shorter tails. [Mobile and extrachromosomal element functions, Prophage functions] |
| pfam10145 | PhageMin_Tail | 1.85e-55 | 362 | 561 | 1 | 200 | Phage-related minor tail protein. Members of this family are found in putative phage tail tape measure proteins. |
| cd13402 | LT_TF-like | 5.48e-36 | 1751 | 1865 | 2 | 117 | lytic transglycosylase-like domain of tail fiber-like proteins and similar domains. These tail fiber-like proteins are multi-domain proteins that include a lytic transglycosylase (LT) domain. Members of the LT family include the soluble and insoluble membrane-bound LTs in bacteria, the LTs in bacteriophage lambda, and the eukaryotic "goose-type" lysozymes (goose egg-white lysozyme; GEWL). LTs catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. |
| COG3953 | SLT | 1.23e-28 | 1753 | 1894 | 18 | 171 | SLT domain protein [Mobilome: prophages, transposons]. |
| cd12797 | M23_peptidase | 3.94e-26 | 1607 | 1692 | 1 | 85 | M23 family metallopeptidase, also known as beta-lytic metallopeptidase, and similar proteins. This model describes the metallopeptidase M23 family, which includes beta-lytic metallopeptidase and lysostaphin. Members of this family are zinc endopeptidases that lyse bacterial cell wall peptidoglycans; they cleave either the N-acylmuramoyl-Ala bond between the cell wall peptidoglycan and the cross-linking peptide (e.g. beta-lytic endopeptidase) or a bond within the cross-linking peptide (e.g. stapholysin, and lysostaphin). Beta-lytic metallopeptidase, formerly known as beta-lytic protease, has a preference for cleavage of Gly-X bonds and favors hydrophobic or apolar residues on either side. It inhibits growth of sensitive organisms and may potentially serve as an antimicrobial agent. Lysostaphin, produced by Staphylococcus genus, cleaves pentaglycine cross-bridges of cell wall peptidoglycan, acting as autolysins to maintain cell wall metabolism or as toxins and weapons against competing strains. Staphylolysin (also known as LasA) is implicated in a range of processes related to Pseudomonas virulence, including stimulating shedding of the ectodomain of cell surface heparan sulphate proteoglycan syndecan-1, and elastin degradation in connective tissue. Its active site is less constricted and contains a five-coordinate zinc ion with trigonal bipyramidal geometry and two metal-bound water molecules, possibly contributing to its activity against a wider range of substrates than those used by related lytic enzymes, consistent with its multiple roles in Pseudomonas virulence. The family includes members that do not appear to have the conserved zinc-binding site and might be lipoproteins lacking proteolytic activity. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRN90722.1 | 0.0 | 1 | 1955 | 1 | 1972 |
| QYG31585.1 | 0.0 | 1 | 1955 | 1 | 1972 |
| ADV05786.1 | 2.33e-294 | 3 | 1953 | 2 | 1861 |
| CDL65134.2 | 7.02e-290 | 24 | 1952 | 1 | 2103 |
| ANQ88262.1 | 3.73e-285 | 3 | 1953 | 2 | 1861 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2B44_A | 2.82e-14 | 1605 | 1713 | 25 | 131 | TruncatedS. aureus LytM, P 32 2 1 crystal form [Staphylococcus aureus],2B44_B Truncated S. aureus LytM, P 32 2 1 crystal form [Staphylococcus aureus] |
| 2B0P_A | 2.90e-14 | 1605 | 1713 | 26 | 132 | truncatedS. aureus LytM, P212121 crystal form [Staphylococcus aureus],2B0P_B truncated S. aureus LytM, P212121 crystal form [Staphylococcus aureus],2B13_A Truncated S. aureus LytM, P41 crystal form [Staphylococcus aureus],2B13_B Truncated S. aureus LytM, P41 crystal form [Staphylococcus aureus],4ZYB_A Chain A, Glycyl-glycine endopeptidase LytM [Staphylococcus aureus subsp. aureus NCTC 8325],4ZYB_B Chain B, Glycyl-glycine endopeptidase LytM [Staphylococcus aureus subsp. aureus NCTC 8325],4ZYB_C Chain C, Glycyl-glycine endopeptidase LytM [Staphylococcus aureus subsp. aureus NCTC 8325],4ZYB_D Chain D, Glycyl-glycine endopeptidase LytM [Staphylococcus aureus subsp. aureus NCTC 8325] |
| 5KQB_A | 1.33e-13 | 1605 | 1709 | 27 | 131 | Identificationand structural characterization of LytU [Staphylococcus aureus],5KQC_A Identification and structural characterization of LytU [Staphylococcus aureus] |
| 6UE4_A | 5.97e-13 | 1556 | 1692 | 229 | 350 | ShyAEndopeptidase from Vibrio cholerae (Closed form) [Vibrio cholerae O1 biovar El Tor str. N16961],6UE4_B ShyA Endopeptidase from Vibrio cholerae (Closed form) [Vibrio cholerae O1 biovar El Tor str. N16961] |
| 6U2A_A | 6.09e-13 | 1556 | 1692 | 226 | 347 | ShyAendopeptidase from Vibrio cholera (open form) [Vibrio cholerae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| E7DNB6 | 4.46e-64 | 275 | 823 | 13 | 514 | Tape measure protein OS=Pneumococcus phage Dp-1 OX=59241 GN=TMP PE=4 SV=1 |
| O64314 | 2.80e-23 | 299 | 661 | 169 | 541 | Probable tape measure protein OS=Escherichia phage P2 OX=10679 GN=T PE=3 SV=1 |
| P45931 | 2.07e-17 | 1753 | 1880 | 1367 | 1502 | Uncharacterized protein YqbO OS=Bacillus subtilis (strain 168) OX=224308 GN=yqbO PE=1 SV=2 |
| P54334 | 6.48e-16 | 1753 | 1880 | 1115 | 1250 | Phage-like element PBSX protein XkdO OS=Bacillus subtilis (strain 168) OX=224308 GN=xkdO PE=4 SV=2 |
| P44693 | 2.10e-12 | 1588 | 1691 | 333 | 432 | Uncharacterized metalloprotease HI_0409 OS=Haemophilus influenzae (strain ATCC 51907 / DSM 11121 / KW20 / Rd) OX=71421 GN=HI_0409 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
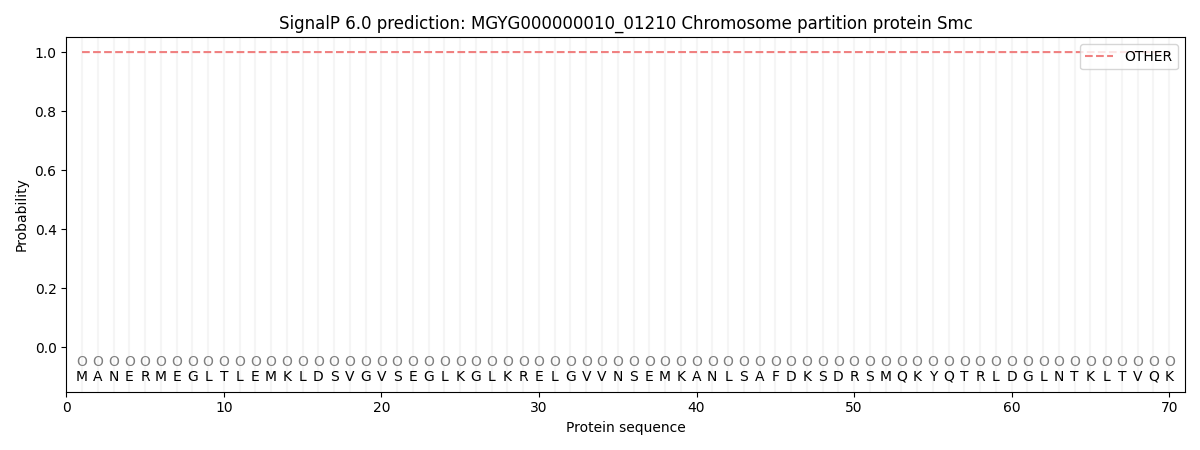
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000066 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |