You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000240_00170
You are here: Home > Sequence: MGYG000000240_00170
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
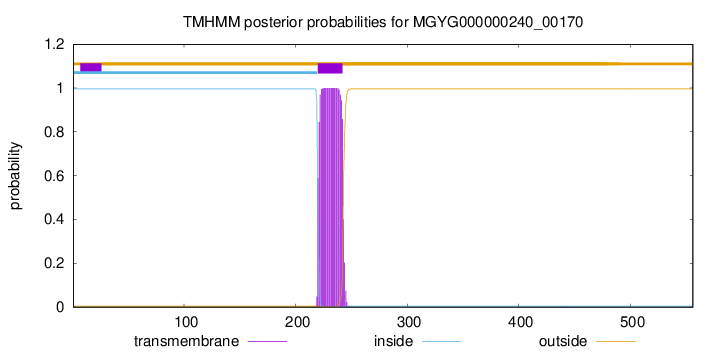
TMHMM annotations
Basic Information help
| Species | Amedibacterium intestinale | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Erysipelotrichales; Erysipelotrichaceae; Amedibacterium; Amedibacterium intestinale | |||||||||||
| CAZyme ID | MGYG000000240_00170 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 14405; End: 16075 Strand: + | |||||||||||
Full Sequence Download help
| MSKNIKTRTV LKDVKTIDKA KVASEHIHHA TIKSKEQYEK HTSQKENTPH DYAVNKTSEK | 60 |
| VPRYTKNTVI NARVNEKRYR NHRNEKNTIK TLQKEYQNNV SSVMQQNHQI KAKTHKTRSV | 120 |
| FTKNRTIKTR RINKKDIKTT DSKGIKQFIK APKKDRKVSV RIPSQNKNVA KQYAIQSMKK | 180 |
| SKESASAMKQ VAMKSSNYAK KISKATTSAL KKMIESAKEV YLFLSAIGSV ACLFILVIAL | 240 |
| VGGIFMSRGN SSSGNAQLSQ EVIAYTPLIQ KYADEFEIPL YVNAIQAVMM QESGGKGNDP | 300 |
| MQSSECAFNK KYPNTPNGIT DPEYSIEVGI ENFADCIKRA KCKDPFDIEN LSLTWQGYNF | 360 |
| GNGYIEWAVK NFGGYSQGNA QVFSEEQAAK HGWSSYGDPE YVPHVMRYYQ FAQLGTGNSQ | 420 |
| LVNVALTQLG NKGGIPYWSW WGYSSRVEWC AIFVSWCSEQ CGMLQDGSMP KFENVTVGMN | 480 |
| WFKERNQWLP RGTVPSEGMI IFFDWNNDNH CDHVGMVEKA DNGIVYTVEG NSNDEVRRNT | 540 |
| YAVNSNVIMG YGTIKK | 556 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd16891 | CwlT-like | 7.94e-58 | 265 | 410 | 1 | 151 | CwlT-like N-terminal lysozyme domain and similar domains. CwlT is a bifunctional cell wall hydrolase containing an N-terminal lysozyme domain and a C-terminal NlpC/P60 endopeptidase domain (gamma-d-D-glutamyl-L-diamino acid endopeptidase), and has been implicated in the spread of transposons. Proteins similar to this family include the soluble and insoluble membrane-bound LTs in bacteria, the LTs in bacteriophage lambda, as well as the eukaryotic "goose-type" lysozymes (goose egg-white lysozyme; GEWL). |
| pfam13702 | Lysozyme_like | 2.00e-51 | 259 | 409 | 1 | 165 | Lysozyme-like. |
| pfam05257 | CHAP | 5.07e-11 | 450 | 531 | 10 | 83 | CHAP domain. This domain corresponds to an amidase function. Many of these proteins are involved in cell wall metabolism of bacteria. This domain is found at the N-terminus of Escherichia coli gss, where it functions as a glutathionylspermidine amidase EC:3.5.1.78. This domain is found to be the catalytic domain of PlyCA. CHAP is the amidase domain of bifunctional Escherichia coli glutathionylspermidine synthetase/amidase, and it catalyzes the hydrolysis of Gsp (glutathionylspermidine) into glutathione and spermidine. |
| NF037998 | RND_1 | 3.49e-04 | 70 | 179 | 1054 | 1149 | protein translocase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNM10615.1 | 0.0 | 1 | 556 | 1 | 556 |
| BBK22824.1 | 2.91e-309 | 1 | 556 | 1 | 550 |
| ANU70499.1 | 3.71e-174 | 1 | 554 | 1 | 570 |
| ASU17078.1 | 3.71e-174 | 1 | 554 | 1 | 570 |
| QQR25626.1 | 3.71e-174 | 1 | 554 | 1 | 570 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4HPE_A | 2.65e-15 | 248 | 516 | 8 | 272 | ChainA, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_B Chain B, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_C Chain C, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_D Chain D, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_E Chain E, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_F Chain F, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630] |
| 4FDY_A | 5.66e-13 | 254 | 437 | 18 | 206 | ChainA, Similar to lipoprotein, NLP/P60 family [Staphylococcus aureus subsp. aureus Mu50],4FDY_B Chain B, Similar to lipoprotein, NLP/P60 family [Staphylococcus aureus subsp. aureus Mu50] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P96645 | 4.34e-31 | 241 | 463 | 36 | 250 | Probable endopeptidase YddH OS=Bacillus subtilis (strain 168) OX=224308 GN=yddH PE=3 SV=1 |
| O34636 | 4.09e-15 | 265 | 390 | 55 | 171 | Uncharacterized membrane protein YocA OS=Bacillus subtilis (strain 168) OX=224308 GN=yocA PE=4 SV=1 |
| Q5XC63 | 2.04e-07 | 260 | 389 | 28 | 148 | Pneumococcal vaccine antigen A homolog OS=Streptococcus pyogenes serotype M6 (strain ATCC BAA-946 / MGAS10394) OX=286636 GN=pvaA PE=3 SV=1 |
| Q8P120 | 2.04e-07 | 260 | 389 | 28 | 148 | Pneumococcal vaccine antigen A homolog OS=Streptococcus pyogenes serotype M18 (strain MGAS8232) OX=186103 GN=pvaA PE=3 SV=1 |
| Q8DPY9 | 7.30e-07 | 240 | 389 | 9 | 149 | Pneumococcal vaccine antigen A OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=pvaA PE=4 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
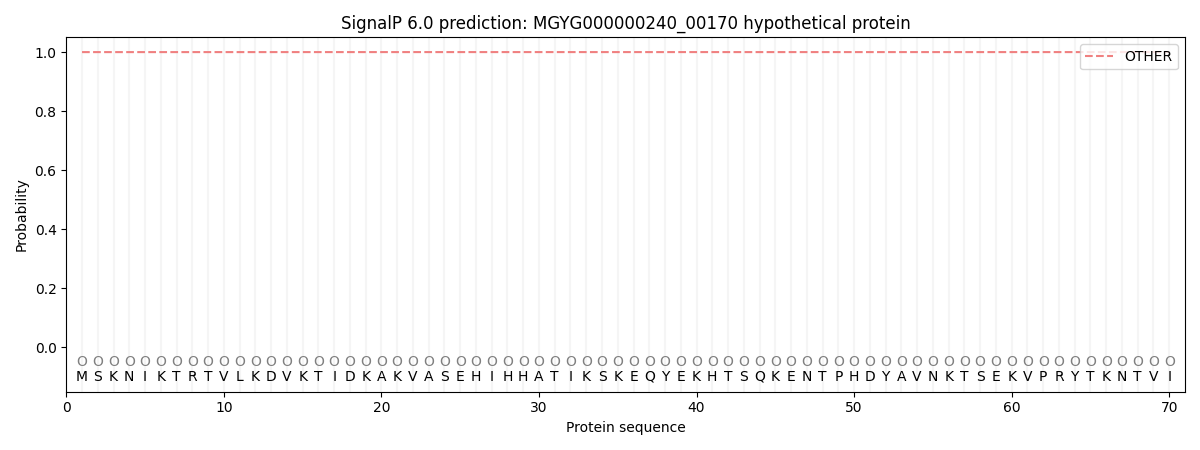
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000034 | 0.000002 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |