You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002301_00590
You are here: Home > Sequence: MGYG000002301_00590
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Staphylococcus warneri | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Staphylococcales; Staphylococcaceae; Staphylococcus; Staphylococcus warneri | |||||||||||
| CAZyme ID | MGYG000002301_00590 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 561029; End: 566623 Strand: + | |||||||||||
Full Sequence Download help
| MADFNLGAEV SLDVDPLKAS KQTLEHNLKD INKSLRSQRA EFKKNELSAE QLAEREKSLG | 60 |
| RAVKLQEGLL ERRKKDLQSV RDEMSKSNVV TDAQRTKLQS ASRAVQQAEN QLSNYNRELK | 120 |
| ETEVAYKQFN RSTDQVKNSL GELKNRAKSS EIAFKQSTRS VDSYKDHLTT MNYTISKSKG | 180 |
| NIDLLKKNLR EVSNAHGSAS RQADKLRNDI LKESIAMQIA QGRADELADE LKEVEREQRK | 240 |
| VALAGTMMIA GFTGARASVD RIATTLRSFG ELTQGIVGEV MATQFANLVP IMGSVVSAGA | 300 |
| GIGGMLTSLA GGAIGLGGAF GIGMGAINAF AGQATYSLKK LEDGELRVTN EVRSYQSALN | 360 |
| GLKSSWDGLI AQNQSAIFNT LTNGIKTANY ALRTLNPFLT TTASQIENMS GRMHTWVTSS | 420 |
| QNARTAFSIL NNQGTKAFGN LLNGAYHFFD GTVAVFNKLS PLFVWASKGF ENMGLSFRRW | 480 |
| ANSVEGSNAI NSFVQYTKTN LPIVGRIFAN VFSGLFNLFG AFAGHSHNVL LGIESVTEGF | 540 |
| KQWSAQLKKS DGFQQFINYL EKNGPKVWSI VKNISMALWG LIKGMAPVAS VTLSLTAAVT | 600 |
| QWLASMTNAH PWIGKLLGST VALTGAMMLF LKPIFLVKGA LTGMRGALLA VTGAETLLGK | 660 |
| QGAFATLGMK RQAVQARISA IATRAWSVAT KAAAFATRGL GLAIRFMSGP IGWVITGIGL | 720 |
| LVGAIVYLWK NNETFRNFVI GAWTAIKNTA ISVFGFLKPY IVGIWNGIKT ASLVVWNLMK | 780 |
| TSATLTWNAI KFAIQNPIQA LKNALSFIWN GIKAGALFAW NSIKVGVMAI IGGWLALVRA | 840 |
| NFAMWRTIIT GVWNGIKYVS ITIWNGIKNG VMAIIRGWIG LVRASFNGLR GFFSAMWNGI | 900 |
| KSVSIAIWNG IKNGVIAIVR ALVGNAKRNI STLRSFISGV WNAIKGISIR VWNAIKNGVI | 960 |
| SAIRGLSSGV RKIIGTLRGW IISAWNYIKN KVINLAKALS NGVKRAFNSL WGAVKKIFGA | 1020 |
| IRNFAVKTWT YIKNKVISLA KGLYSGVKRA FTGTWNFVKR VFNNIKNFSV KVWNYIKNKV | 1080 |
| VSLAKGLYNG VKRNFTVTWN ITKSIFGKLR GWLTKTWKNI KNSVVSHARG LWSGVKGTWS | 1140 |
| RLKSGTSSTF GRVKSDTISK WKSMKSSVTG LAKGLWSSVR NTFRNMASGL KTLIGRIKSH | 1200 |
| INNMVGGVKG GLNKLIGGVN WVAGKLGMDK LPTLKLHTGT THTQKYVTNG KINQDTFATV | 1260 |
| GDKGRGNGPG GFRHEMIRYP NGRTAITPNR DTTAFLPKGS QVYNGAQTHS MLNNHPAFAT | 1320 |
| GTLPKFASGT MFNLLGGGKK PKKHKHKDAN DSDESLGSKL GNMWGGVKNA TSKAVSTGVG | 1380 |
| WAKAAGKTAS KVIGDVLDYI EKPSKLVEKV FDIFGVGVKT FGIPSGAELP FNMMKGMFGK | 1440 |
| LKKGAIDKVK NWLEESGGGD GGYIKYLDNI TTPYSPNGPP PGYAFSWPHP GIDLPYKYEP | 1500 |
| VYSTISGTAR GKEMPGGFGH YIEVIGGALK VIYGHLSKWN IKQGQKVHPG TKLGVSGNTG | 1560 |
| ASTGPHLHYE MHRNGKPIDP VKWLKSHNGG GKSGGSRAAS AWRPEVIKAL RANHLPTSGA | 1620 |
| YVNAWIRQID SESSGNAGAH QGIHDVNSGG NEAQGLVQVT PATFRAFKMA GHGNILNGLD | 1680 |
| NLMAGIHYAK SRYGSSMLSV IGHGHGYAKG TNNARKGFAN IFEEGGEIMQ MHGGETVIPN | 1740 |
| DVSIEAFKQI ANSDIFARTQ SAVYEAISRY ADEIRQDKAQ KQQEQYRKDM EYQALKEQNN | 1800 |
| KLTSLVDKMD TIISSLLNIE DSSERTANKK TVIDKFSHEK EVNSIVDKRE RNKVRTSKFK | 1860 |
| PRLT | 1864 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd13402 | LT_TF-like | 2.90e-35 | 1622 | 1737 | 1 | 117 | lytic transglycosylase-like domain of tail fiber-like proteins and similar domains. These tail fiber-like proteins are multi-domain proteins that include a lytic transglycosylase (LT) domain. Members of the LT family include the soluble and insoluble membrane-bound LTs in bacteria, the LTs in bacteriophage lambda, and the eukaryotic "goose-type" lysozymes (goose egg-white lysozyme; GEWL). LTs catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. |
| COG5412 | COG5412 | 1.32e-29 | 898 | 1313 | 81 | 501 | Phage-related protein [Mobilome: prophages, transposons]. |
| pfam01551 | Peptidase_M23 | 6.14e-27 | 1488 | 1580 | 2 | 96 | Peptidase family M23. Members of this family are zinc metallopeptidases with a range of specificities. The peptidase family M23 is included in this family, these are Gly-Gly endopeptidases. Peptidase family M23 are also endopeptidases. This family also includes some bacterial lipoproteins for which no proteolytic activity has been demonstrated. This family also includes leukocyte cell-derived chemotaxin 2 (LECT2) proteins. LECT2 is a liver-specific protein which is thought to be linked to hepatocyte growth although the exact function of this protein is unknown. |
| COG5412 | COG5412 | 3.69e-22 | 672 | 1054 | 153 | 529 | Phage-related protein [Mobilome: prophages, transposons]. |
| COG3953 | SLT | 6.03e-21 | 1627 | 1859 | 20 | 232 | SLT domain protein [Mobilome: prophages, transposons]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AGZ26280.1 | 0.0 | 175 | 1861 | 231 | 1925 |
| AID01516.1 | 0.0 | 1 | 1829 | 1 | 1715 |
| AGW43750.1 | 0.0 | 1 | 1830 | 1 | 1784 |
| QXV71939.1 | 0.0 | 1 | 1833 | 1 | 1807 |
| QSF50925.1 | 2.59e-226 | 226 | 1861 | 288 | 1942 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6V8I_AF | 4.31e-73 | 259 | 747 | 246 | 734 | ChainAF, Tape Measure Protein, gp57 [Dubowvirus dv80alpha],6V8I_BF Chain BF, Tape Measure Protein, gp57 [Dubowvirus dv80alpha],6V8I_CF Chain CF, Tape Measure Protein, gp57 [Dubowvirus dv80alpha] |
| 5J1L_A | 1.85e-13 | 1489 | 1585 | 65 | 165 | Crystalstructure of Csd1-Csd2 dimer I [Helicobacter pylori 26695],5J1L_C Crystal structure of Csd1-Csd2 dimer I [Helicobacter pylori 26695],5J1M_A Crystal structure of Csd1-Csd2 dimer II [Helicobacter pylori 26695],5J1M_C Crystal structure of Csd1-Csd2 dimer II [Helicobacter pylori 26695] |
| 4RNY_A | 1.90e-11 | 1464 | 1586 | 195 | 319 | Structureof Helicobacter pylori Csd3 from the orthorhombic crystal [Helicobacter pylori 26695],4RNY_B Structure of Helicobacter pylori Csd3 from the orthorhombic crystal [Helicobacter pylori 26695],4RNZ_A Structure of Helicobacter pylori Csd3 from the hexagonal crystal [Helicobacter pylori 26695] |
| 6UE4_A | 2.17e-10 | 1488 | 1582 | 265 | 361 | ShyAEndopeptidase from Vibrio cholerae (Closed form) [Vibrio cholerae O1 biovar El Tor str. N16961],6UE4_B ShyA Endopeptidase from Vibrio cholerae (Closed form) [Vibrio cholerae O1 biovar El Tor str. N16961] |
| 6U2A_A | 2.20e-10 | 1488 | 1582 | 262 | 358 | ShyAendopeptidase from Vibrio cholera (open form) [Vibrio cholerae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q0PDK7 | 4.95e-28 | 286 | 627 | 84 | 447 | Tail tape measure protein gp18 OS=Bacillus phage SPP1 OX=10724 PE=4 SV=1 |
| P44693 | 9.09e-14 | 1470 | 1581 | 329 | 443 | Uncharacterized metalloprotease HI_0409 OS=Haemophilus influenzae (strain ATCC 51907 / DSM 11121 / KW20 / Rd) OX=71421 GN=HI_0409 PE=3 SV=1 |
| P0AFS9 | 4.63e-10 | 1470 | 1611 | 295 | 438 | Murein DD-endopeptidase MepM OS=Escherichia coli (strain K12) OX=83333 GN=mepM PE=1 SV=1 |
| P0AFT0 | 4.63e-10 | 1470 | 1611 | 295 | 438 | Murein DD-endopeptidase MepM OS=Escherichia coli O6:H1 (strain CFT073 / ATCC 700928 / UPEC) OX=199310 GN=mepM PE=3 SV=1 |
| P0AFT1 | 4.63e-10 | 1470 | 1611 | 295 | 438 | Murein DD-endopeptidase MepM OS=Shigella flexneri OX=623 GN=mepM PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
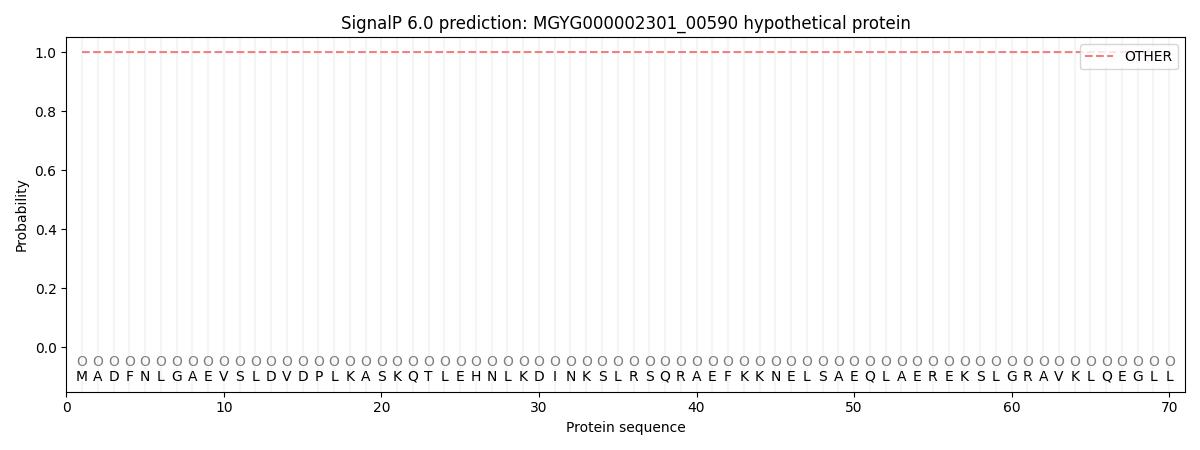
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000082 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
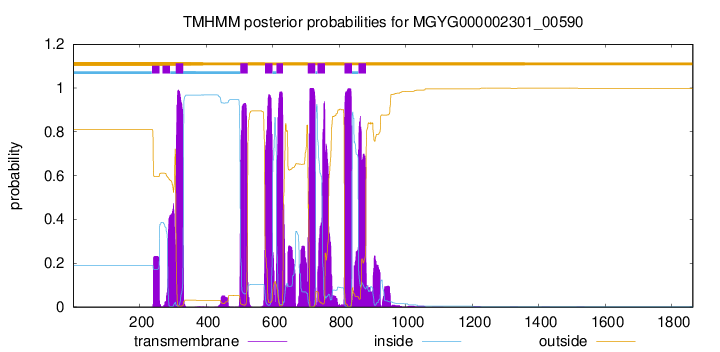
| start | end |
|---|---|
| 309 | 331 |
| 503 | 525 |
| 577 | 599 |
| 612 | 631 |
| 706 | 728 |
| 735 | 757 |
| 816 | 838 |
| 858 | 880 |
