You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004123_00721
You are here: Home > Sequence: MGYG000004123_00721
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Peptostreptococcales; Anaerovoracaceae; QAKD01; | |||||||||||
| CAZyme ID | MGYG000004123_00721 | |||||||||||
| CAZy Family | GH26 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 74460; End: 76007 Strand: + | |||||||||||
Full Sequence Download help
| MKKIIGILTA VIMVFSVPVE ASEASLRGQP EELYRLISAG NYDLFTNYVD GYEMFVDRGM | 60 |
| SVDTDYFGVA AVLENETKRI EIYKQNLSGI SKSGYINYSN QFLKNTADHY GIVQQTEEIG | 120 |
| GRSIQITAWE RNKLARVSND KNYYVCLEIQ EGSNVYTIFV KSAVPIAENG GYRYLAENFQ | 180 |
| LIRSTKSGFI RMSETIPAEE KGWNAETYEF YQKYFNDEAS LSWGIFEPDV DYTGYDTLHG | 240 |
| YERTLDYIFP VLLSYTEVIN TAPEVVKNRL EDAYENGKVL ELTLQTAHAS NNMIYNLLNG | 300 |
| QYDSYLYEYA SVISEFGHPV LFRLFNEMNG DWCPYSSYNT SKDTMLFKEA YWYIYSIFDS | 360 |
| AGANENTIWI WNPNSESYPD FNWNHALMYY PGDEYVDVVG LTAYNTGTYY SGEKWQSFSE | 420 |
| LYDNLYSDYC QWFSQPFMIT EFASSSIGGD KAAWIDDMFL RMQNFSRIKI AIWWDGCDWD | 480 |
| ANGNIARPYF LDETPDTLEA FRKGIRSSWN VDVYA | 515 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH26 | 298 | 421 | 2.3e-25 | 0.37293729372937295 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02156 | Glyco_hydro_26 | 1.89e-13 | 295 | 410 | 129 | 242 | Glycosyl hydrolase family 26. |
| COG4124 | ManB2 | 1.23e-11 | 297 | 441 | 155 | 294 | Beta-mannanase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QAT42469.1 | 2.97e-215 | 1 | 515 | 4 | 520 |
| QHI71412.1 | 4.38e-214 | 1 | 515 | 1 | 517 |
| QIB68979.1 | 5.48e-210 | 1 | 514 | 1 | 516 |
| QOX62356.1 | 9.83e-202 | 5 | 511 | 12 | 519 |
| QUH19438.1 | 1.46e-190 | 35 | 507 | 49 | 525 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2BVD_A | 4.15e-26 | 266 | 505 | 51 | 280 | ChainA, ENDOGLUCANASE H [Acetivibrio thermocellus] |
| 2V3G_A | 4.15e-26 | 266 | 505 | 51 | 280 | ChainA, Endoglucanase H [Acetivibrio thermocellus] |
| 2BV9_A | 4.77e-26 | 266 | 505 | 51 | 280 | ChainA, Endoglucanase H [Acetivibrio thermocellus] |
| 2CIP_A | 1.04e-25 | 266 | 505 | 51 | 280 | ChainA, ENDOGLUCANASE H [Acetivibrio thermocellus] |
| 2VI0_A | 1.04e-25 | 266 | 505 | 51 | 280 | ChainA, Endoglucanase H [Acetivibrio thermocellus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P16218 | 1.04e-23 | 266 | 505 | 73 | 302 | Endoglucanase H OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celH PE=1 SV=1 |
| O05512 | 1.65e-07 | 317 | 409 | 183 | 273 | Mannan endo-1,4-beta-mannosidase OS=Bacillus subtilis (strain 168) OX=224308 GN=gmuG PE=1 SV=2 |
| P55278 | 2.90e-07 | 317 | 475 | 181 | 323 | Mannan endo-1,4-beta-mannosidase OS=Bacillus subtilis OX=1423 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
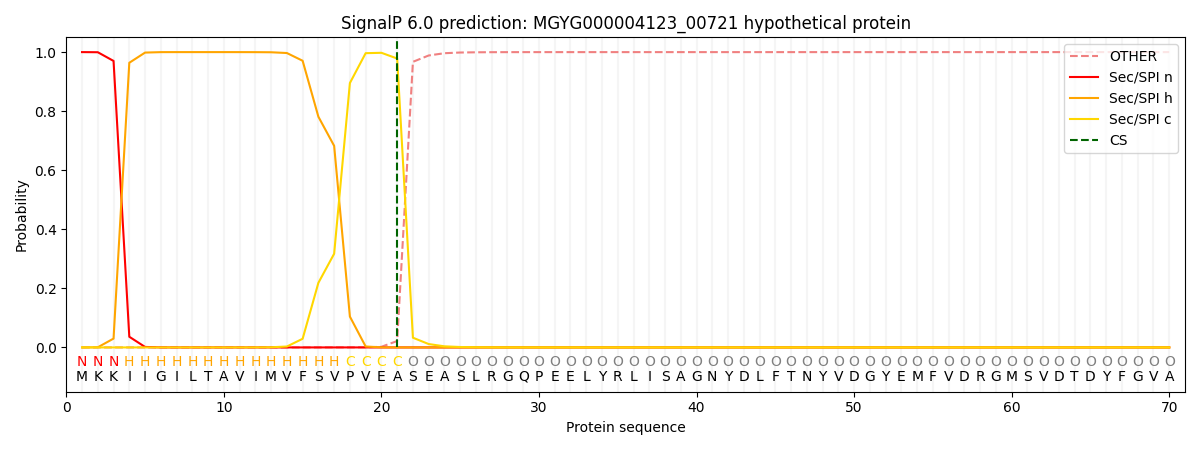
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000278 | 0.998921 | 0.000200 | 0.000203 | 0.000196 | 0.000173 |
