You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000013_02452
You are here: Home > Sequence: MGYG000000013_02452
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides sp902362375 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides sp902362375 | |||||||||||
| CAZyme ID | MGYG000000013_02452 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Xylan 1,4-beta-xylosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 480295; End: 482874 Strand: + | |||||||||||
Full Sequence Download help
| MKHLYLSILL LLSLAACTPQ PPQYDFQDPD KPIEARVENL LSLLTLEEKV GQMMHEAPAI | 60 |
| DRLGIPSYNW WNECLHGVAR SGQATVFPQA IGMAATFDNE LIFNIATAIS DEARAKHHDF | 120 |
| KKRGWHDYYQ GLTYWTPNIN IFRDPRWGRG METYGEDPLL TARMGVAFIK GLQGDDPNHF | 180 |
| KLIATPKHYV VHSGPEYNRH SFDAVADMYD FYMTYLPAFD RAIKEANAQS VMCAYNRTNG | 240 |
| EVCCGSDELL NMILRHELKF NGYIVSDCGA ISNFYNEHEH HVVNTPAEAA AKAVRAGTDL | 300 |
| NCGETYHALI DAVKQGLLKD CDIDVSLGRL LKARFQLGMF DPDERVAYSR IPLTVVECKE | 360 |
| HVELAREAAR KSMVLLKNDK SILPLSKEKK RIAVIGPNAD NVETLYANYN GFSKNPVTAL | 420 |
| QGIKNKLQDS EILYAQGCNL ANGLPYLTPI PTDYLFTDAS CTQHGVTGHY FDNDTLAGNA | 480 |
| VLTRRDPNID FNWWHEAPLK ELDPHNFSVR WTGYLKAPAT GDYYLGYHSA YLSMWVDNQL | 540 |
| IGSTHDVHHP TKSYKKLTLE AGKVYELKLE LKKHRGMAFE RLLWQRPDNH LQEEAIATAT | 600 |
| QSDMIILCMG LSPLLEGEEM NVDLEGFYKG DRTTLDLPAP QLALMKELKK LNKPMVLVLM | 660 |
| NGSAVSVNWE KENMNAILEA WYPGQEGGNA IADILFGDYT PGGKLPVTFY KSVNDLPDFE | 720 |
| NYSMKGHTYR YFEKETLWNF GYGLSYTTFQ YNDLTLMDAT PGQMKVKVNV SNTGKMDGDD | 780 |
| VVQVYVRKVS PRTEHHPRYT LAGFKRISLK KEKRKEVVLA LDDMAFMELD DNGQKVIGKG | 840 |
| SYELFVGADS KEIVKFKID | 859 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 60 | 302 | 7.9e-70 | 0.9768518518518519 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PLN03080 | PLN03080 | 3.40e-138 | 19 | 847 | 35 | 769 | Probable beta-xylosidase; Provisional |
| PRK15098 | PRK15098 | 5.79e-128 | 16 | 853 | 14 | 757 | beta-glucosidase BglX. |
| COG1472 | BglX | 2.01e-74 | 52 | 440 | 47 | 397 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| pfam01915 | Glyco_hydro_3_C | 1.35e-56 | 373 | 746 | 1 | 216 | Glycosyl hydrolase family 3 C-terminal domain. This domain is involved in catalysis and may be involved in binding beta-glucan. This domain is found associated with pfam00933. |
| pfam00933 | Glyco_hydro_3 | 3.67e-36 | 62 | 333 | 63 | 316 | Glycosyl hydrolase family 3 N terminal domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBD44113.1 | 0.0 | 1 | 847 | 1 | 852 |
| SCM55255.1 | 2.30e-317 | 1 | 847 | 1 | 852 |
| QIK60870.1 | 6.38e-313 | 5 | 853 | 10 | 862 |
| AZQ59364.1 | 2.48e-311 | 8 | 847 | 12 | 856 |
| QIK55454.1 | 2.39e-310 | 5 | 853 | 10 | 862 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7VC7_A | 4.34e-88 | 35 | 858 | 24 | 729 | ChainA, xylan 1,4-beta-xylosidase [Phanerodontia chrysosporium] |
| 7VC6_A | 4.34e-88 | 35 | 858 | 24 | 729 | ChainA, xylan 1,4-beta-xylosidase [Phanerodontia chrysosporium] |
| 6Q7I_A | 2.54e-83 | 36 | 858 | 52 | 751 | GH3exo-beta-xylosidase (XlnD) [Aspergillus nidulans FGSC A4],6Q7I_B GH3 exo-beta-xylosidase (XlnD) [Aspergillus nidulans FGSC A4],6Q7J_A Chain A, Exo-1,4-beta-xylosidase xlnD [Aspergillus nidulans FGSC A4],6Q7J_B Chain B, Exo-1,4-beta-xylosidase xlnD [Aspergillus nidulans FGSC A4] |
| 5A7M_A | 7.73e-79 | 36 | 858 | 52 | 753 | ChainA, BETA-XYLOSIDASE [Trichoderma reesei],5A7M_B Chain B, BETA-XYLOSIDASE [Trichoderma reesei] |
| 5AE6_A | 7.88e-79 | 36 | 858 | 52 | 753 | ChainA, BETA-XYLOSIDASE [Trichoderma reesei],5AE6_B Chain B, BETA-XYLOSIDASE [Trichoderma reesei] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| D5EY15 | 1.01e-197 | 1 | 858 | 1 | 859 | Xylan 1,4-beta-xylosidase OS=Prevotella ruminicola (strain ATCC 19189 / JCM 8958 / 23) OX=264731 GN=xyl3A PE=1 SV=1 |
| Q94KD8 | 1.72e-104 | 32 | 853 | 50 | 756 | Probable beta-D-xylosidase 2 OS=Arabidopsis thaliana OX=3702 GN=BXL2 PE=2 SV=1 |
| Q9SGZ5 | 8.88e-104 | 16 | 847 | 25 | 757 | Probable beta-D-xylosidase 7 OS=Arabidopsis thaliana OX=3702 GN=BXL7 PE=2 SV=2 |
| B0Y0I4 | 3.70e-103 | 28 | 846 | 44 | 740 | Probable exo-1,4-beta-xylosidase bxlB OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=bxlB PE=3 SV=1 |
| Q4WFI6 | 3.70e-103 | 28 | 846 | 44 | 740 | Probable exo-1,4-beta-xylosidase bxlB OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=bxlB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
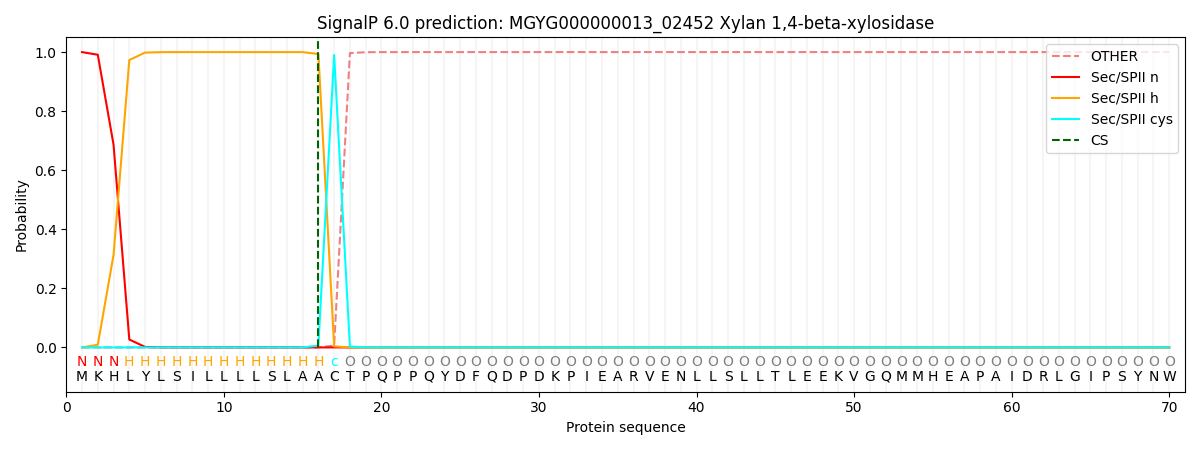
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000015 | 1.000024 | 0.000000 | 0.000000 | 0.000000 |
