You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000013_04627
You are here: Home > Sequence: MGYG000000013_04627
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides sp902362375 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides sp902362375 | |||||||||||
| CAZyme ID | MGYG000000013_04627 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Beta-glucosidase BoGH3B | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 41091; End: 43385 Strand: + | |||||||||||
Full Sequence Download help
| MKLRNIILMS ASIAMTACSK QAVPTAIPED AKIEQQVEEL LSKMDLDAKI GQMTELAIDV | 60 |
| LGDIVNGEFQ LDETKLHKAI AEYKVGSFLN APGPVAQSPE KWNEIISRIQ ELSMKEIGIP | 120 |
| CIYGLDQNHG TTYTLGGTLF PQNINMGAAF NPELAYEAAR VTAYETRASN CPWTYSPTVD | 180 |
| MARDPRWPRV WENYGEDCLV NAIMGSNAVR GFQGDDPNHI PADRIATSVK HYMGYSIPRT | 240 |
| GKDRTPAYIS VSELREKCFA PFKACVEAGA LTIMVNSGSI NGKPVHADHE LLTKWLKEDL | 300 |
| GWDGMLITDW ADINNLYTRE HVAADKKEAI EMAINAGIDM AMEPYDLNYC TLLKELVQEK | 360 |
| KVPMSRIDDA VRRVLRLKFR LGLFDHPNTL LKDYPLFGSK EHALIALHAA EESEVLLKNK | 420 |
| DNILPLPQGK KILVTGPNAN SMRCLNGGWS YSWQGHLTDR FADKYNTIYE AICNKFGTDH | 480 |
| VRLEQGVTYK PEGAYMEENE PEIEKAVAAA RNVDIIIACI GENSYCETPG NLSELAISAN | 540 |
| QSKLVKALAV TEKPIILILN EGRPRIINDL EPLVDAVINV LLPGNYGGDA LANILAGDVN | 600 |
| PSAKMPYTYP RHEAALTTYD YRVSEEMDKM EGAYDYNAVV SVQWPFGYGL SYTTFEYSNF | 660 |
| QTDKSSFSTG DKLHFSIDVT NTGKYAGKEV VMLFSSDLVA SLTPENRRLR AFKKVELQPG | 720 |
| ETQTVTLSIK DSDLAFVNAD GQWVLEQGDF RMQCGNQVLT ITYK | 764 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 115 | 342 | 2.6e-62 | 0.9722222222222222 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK15098 | PRK15098 | 1.03e-141 | 8 | 755 | 4 | 751 | beta-glucosidase BglX. |
| COG1472 | BglX | 1.65e-79 | 44 | 451 | 1 | 373 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| pfam00933 | Glyco_hydro_3 | 4.17e-62 | 46 | 377 | 2 | 316 | Glycosyl hydrolase family 3 N terminal domain. |
| PLN03080 | PLN03080 | 1.42e-61 | 108 | 762 | 65 | 776 | Probable beta-xylosidase; Provisional |
| pfam01915 | Glyco_hydro_3_C | 2.97e-50 | 415 | 652 | 2 | 216 | Glycosyl hydrolase family 3 C-terminal domain. This domain is involved in catalysis and may be involved in binding beta-glucan. This domain is found associated with pfam00933. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNL39941.1 | 0.0 | 1 | 764 | 1 | 764 |
| QUT31664.1 | 0.0 | 1 | 764 | 1 | 764 |
| QGT72556.1 | 0.0 | 1 | 764 | 1 | 764 |
| QDM10585.1 | 0.0 | 1 | 764 | 1 | 764 |
| QUT79171.1 | 0.0 | 1 | 764 | 1 | 764 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5JP0_A | 0.0 | 21 | 761 | 1 | 746 | Bacteroidesovatus Xyloglucan PUL GH3B with bound glucose [Bacteroides ovatus],5JP0_B Bacteroides ovatus Xyloglucan PUL GH3B with bound glucose [Bacteroides ovatus] |
| 5Z87_A | 1.20e-107 | 112 | 755 | 116 | 771 | ChainA, EmGH1 [Aurantiacibacter marinus],5Z87_B Chain B, EmGH1 [Aurantiacibacter marinus] |
| 6R5I_A | 6.42e-102 | 34 | 755 | 4 | 719 | ChainA, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5I_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5N_A Chain A, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5N_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1] |
| 6R5O_A | 3.43e-101 | 34 | 755 | 4 | 719 | ChainA, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5O_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5R_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5T_A Chain A, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5T_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5U_A Chain A, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5U_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5V_A Chain A, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5V_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1] |
| 6R5R_A | 3.51e-101 | 34 | 755 | 5 | 720 | ChainA, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A7LXU3 | 0.0 | 21 | 761 | 23 | 768 | Beta-glucosidase BoGH3B OS=Bacteroides ovatus (strain ATCC 8483 / DSM 1896 / JCM 5824 / BCRC 10623 / CCUG 4943 / NCTC 11153) OX=411476 GN=BACOVA_02659 PE=1 SV=1 |
| Q23892 | 3.54e-155 | 37 | 759 | 80 | 816 | Lysosomal beta glucosidase OS=Dictyostelium discoideum OX=44689 GN=gluA PE=1 SV=2 |
| P33363 | 8.41e-106 | 37 | 755 | 38 | 751 | Periplasmic beta-glucosidase OS=Escherichia coli (strain K12) OX=83333 GN=bglX PE=3 SV=2 |
| Q56078 | 8.41e-106 | 37 | 755 | 38 | 751 | Periplasmic beta-glucosidase OS=Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) OX=99287 GN=bglX PE=3 SV=2 |
| T2KMH0 | 1.73e-96 | 16 | 755 | 16 | 707 | Beta-xylosidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22130 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
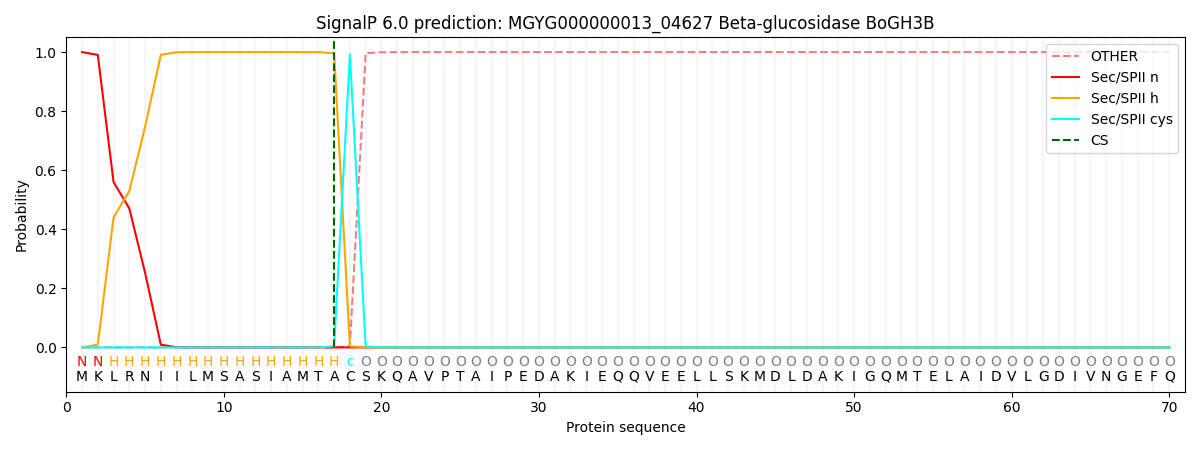
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000042 | 0.000000 | 0.000000 | 0.000000 |
