You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000478_00109
You are here: Home > Sequence: MGYG000000478_00109
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
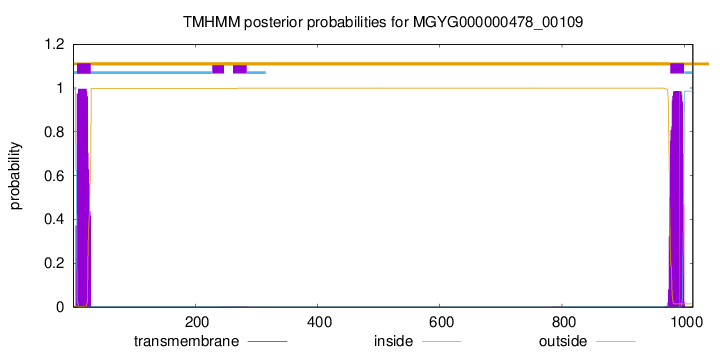
TMHMM annotations
Basic Information help
| Species | UBA4951 sp900540385 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; RFN20; CAG-826; UBA4951; UBA4951 sp900540385 | |||||||||||
| CAZyme ID | MGYG000000478_00109 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 130479; End: 133523 Strand: - | |||||||||||
Full Sequence Download help
| MNKKIANAIV FGVTSAAVIA GLIVGNSMVK RYESEINSVL NPPIIDQEAL NQSASNGQEL | 60 |
| SKKLMQEGAI LLQNDGTLPL DYSATKKVNV FGWRSVDWIH GSDGRNASGR VAPEDGDYTK | 120 |
| NIDLVKALQN YGVETNTRLY NMYRSYSKPM WELMDTRNSH INDMTPLREP NITDLSGSEK | 180 |
| EGSYTNDLLD YCKNFSDTAF VVIGRMAGEG MNCSVNDQVK EGNGAKNDNS RHYLEISTEE | 240 |
| EALLRYCGEN YANVVVFINA ANPFEMGFMK TIPGLDAAMY VGFTGTRAAS ALPKLIYGEV | 300 |
| SPSGRTVDTF PYDLFTNPAN IYLGGQSYLD FNKGYTDAIE NVYLGYKWYE SADQEHYWDK | 360 |
| VNNEYGKGYD GVVYAPFGYG LSYNEYSWNI SKIELKREGK EAVEYEEGVA FTDKDKIEFT | 420 |
| VSVTNNGKVP GQDVVEIYAV APYYGHSEDE AIEKPYVNLA GFAKTNILQP GSTEEITIEV | 480 |
| DPYDFASYDC YDRNNNGFKG YELEHGAYTL SLRSSAHSVK NVTFASKEVA GDFSFNVTQD | 540 |
| IKIGNDPVTG KPVKNLFTGE DAVDMTPIDS QEKDGSYDPG IKWLTRKNFT SLEDLKTSYV | 600 |
| ARRNVTPSAK GQADSVDRIK AWNEATGVDA FGEEIPTEKP TWGASNGLKV ADNGIINEFG | 660 |
| EKLGKDYNDP DWERLLDQLS IEEVVNMIGG YYGSKALPSI GKPALADVDG PSQIKSFVNA | 720 |
| PRGTGYPTMT TIAATWNKKL LYEFGKSFGD DMKGVGIMGD WGFAMDCHRT SFFGRNHESP | 780 |
| SEDMMLAGTL MAEAVRGLGT RGRYNMMKHF ALYGYGGQQI WMTEQGLREN FLKSFRKVVL | 840 |
| DGGCLGVMTT YQGVGGEHSE TTQALLRGVL RGEWDFKGAI TTDYIGHNPY IDTLLRCGGD | 900 |
| FGMGVKLGTI DGVKYDSSSS ARVQHALRDV AHHVTYMWLR ADYYEKQYKA NPTDDNEAFI | 960 |
| SSVSINSWCW WKPLIFTINV TAGTLLTMWG AMVLVSFLIK DEKKEKKLDA KEGK | 1014 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00933 | Glyco_hydro_3 | 4.04e-21 | 673 | 892 | 40 | 263 | Glycosyl hydrolase family 3 N terminal domain. |
| COG1472 | BglX | 2.94e-18 | 723 | 888 | 80 | 253 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| PRK15098 | PRK15098 | 1.49e-15 | 185 | 489 | 465 | 734 | beta-glucosidase BglX. |
| PLN03080 | PLN03080 | 2.28e-10 | 254 | 519 | 532 | 775 | Probable beta-xylosidase; Provisional |
| PRK15098 | PRK15098 | 2.30e-10 | 724 | 898 | 118 | 298 | beta-glucosidase BglX. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QOS39538.1 | 4.12e-215 | 3 | 983 | 2 | 1002 |
| QUC03329.1 | 2.26e-189 | 1 | 952 | 12 | 939 |
| QOY60704.1 | 1.56e-168 | 12 | 941 | 23 | 959 |
| ADK67194.1 | 1.20e-164 | 16 | 941 | 31 | 970 |
| QOS39329.1 | 3.28e-144 | 46 | 1000 | 46 | 972 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5WUG_A | 2.46e-45 | 59 | 884 | 38 | 748 | Expression,characterization and crystal structure of a novel beta-glucosidase from Paenibacillus barengoltzii [Paenibacillus barengoltzii],5WVP_A Expression, characterization and crystal structure of a novel beta-glucosidase from Paenibacillus barengoltzii [Paenibacillus barengoltzii] |
| 2X40_A | 3.24e-23 | 675 | 900 | 7 | 258 | Structureof beta-glucosidase 3B from Thermotoga neapolitana in complex with glycerol [Thermotoga neapolitana DSM 4359],2X41_A Structure of beta-glucosidase 3B from Thermotoga neapolitana in complex with glucose [Thermotoga neapolitana DSM 4359] |
| 2X42_A | 2.97e-22 | 675 | 900 | 7 | 258 | Structureof beta-glucosidase 3B from Thermotoga neapolitana in complex with alpha-D-glucose [Thermotoga neapolitana DSM 4359] |
| 7MS2_A | 6.02e-20 | 231 | 519 | 424 | 660 | ChainA, Thermostable beta-glucosidase B [Acetivibrio thermocellus AD2],7MS2_B Chain B, Thermostable beta-glucosidase B [Acetivibrio thermocellus AD2] |
| 3AC0_A | 3.52e-19 | 671 | 886 | 5 | 228 | Crystalstructure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus],3AC0_B Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus],3AC0_C Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus],3AC0_D Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P16084 | 1.12e-57 | 59 | 904 | 30 | 789 | Beta-glucosidase A OS=Butyrivibrio fibrisolvens OX=831 GN=bglA PE=3 SV=1 |
| P15885 | 1.13e-48 | 59 | 884 | 11 | 697 | Beta-glucosidase OS=Ruminococcus albus OX=1264 PE=3 SV=1 |
| Q5BFG8 | 3.96e-25 | 673 | 961 | 12 | 304 | Beta-glucosidase B OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=bglB PE=1 SV=1 |
| Q9P6J6 | 1.84e-23 | 671 | 886 | 5 | 228 | Putative beta-glucosidase OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPBC1683.04 PE=3 SV=1 |
| Q4WA69 | 3.10e-21 | 669 | 886 | 10 | 235 | Probable beta-glucosidase K OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=bglK PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
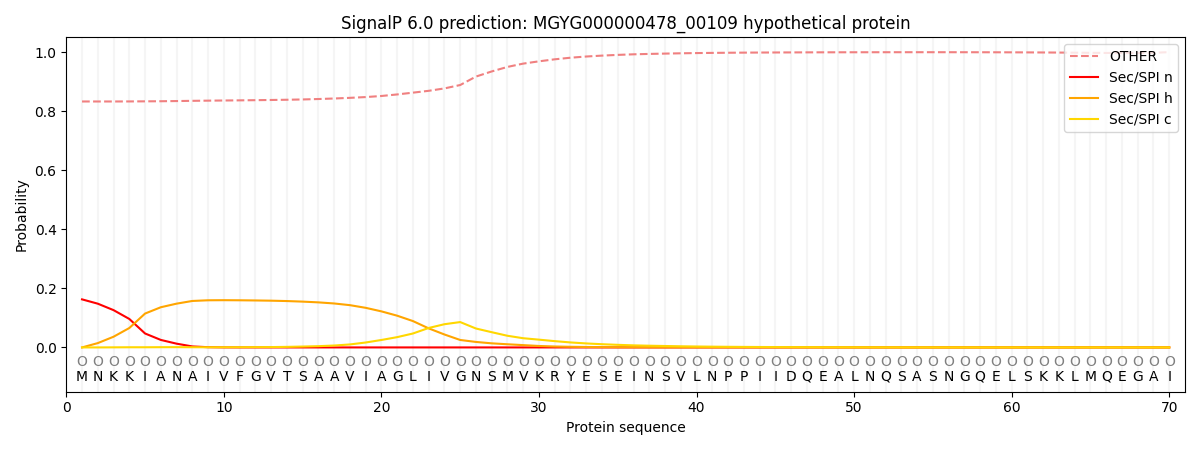
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.842054 | 0.151043 | 0.003716 | 0.000558 | 0.000354 | 0.002282 |