You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001288_00605
You are here: Home > Sequence: MGYG000001288_00605
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Veillonella dispar | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_C; Negativicutes; Veillonellales; Veillonellaceae; Veillonella; Veillonella dispar | |||||||||||
| CAZyme ID | MGYG000001288_00605 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Beta-hexosaminidase A | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 41411; End: 42559 Strand: - | |||||||||||
Full Sequence Download help
| MFRRIVAATM ISALALTTGC GLHNPFTSKA EPVTYESVAQ SELSPEEKVD KLVANMSDAD | 60 |
| KVGQLLMIGI HGKTLNDDAK FMLNEYRVGG IILFDRNMES KDQVKSLIAD INKTGKSAGL | 120 |
| TPLFIGIDQE GGAVARMEDQ LIKVPPAEEL GKEPIEQAIS LAKQSGTELK DLGFNINFAP | 180 |
| VADLGLTYGR SFSTNPDEVV RYASAVGKAY DEAGLWYSYK HFPGIGKTDV DLHADTSVVP | 240 |
| VSKETLLNED TKVFVDLIKQ SKPNTYAIMV SHAMYPQIDP DHPSSLSKAI ITDWLRKDMG | 300 |
| YNGVVVTDDM DMGALAKHYT FGDMAVQSIL AGSDILLVCH EYEHMQEAYN GLMKAVKDGR | 360 |
| ISKERLDESV KRILLMKITK IS | 382 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 121 | 338 | 5.4e-43 | 0.9583333333333334 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1472 | BglX | 3.63e-78 | 56 | 378 | 1 | 313 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| pfam00933 | Glyco_hydro_3 | 6.92e-71 | 60 | 374 | 4 | 314 | Glycosyl hydrolase family 3 N terminal domain. |
| PRK05337 | PRK05337 | 2.23e-56 | 66 | 363 | 4 | 304 | beta-hexosaminidase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBU37286.1 | 3.37e-272 | 1 | 382 | 1 | 382 |
| BBU35398.1 | 6.52e-270 | 1 | 382 | 1 | 382 |
| VEG94458.1 | 6.52e-270 | 1 | 380 | 1 | 380 |
| QQB17095.1 | 3.06e-261 | 1 | 381 | 1 | 381 |
| ACZ25449.1 | 3.06e-261 | 1 | 381 | 1 | 381 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6K5J_A | 1.55e-65 | 61 | 377 | 16 | 337 | Structureof a glycoside hydrolase family 3 beta-N-acetylglucosaminidase from Paenibacillus sp. str. FPU-7 [Paenibacillaceae] |
| 4ZM6_A | 2.94e-42 | 53 | 377 | 3 | 337 | Aunique GCN5-related glucosamine N-acetyltransferase region exist in the fungal multi-domain GH3 beta-N-acetylglucosaminidase [Rhizomucor miehei CAU432],4ZM6_B A unique GCN5-related glucosamine N-acetyltransferase region exist in the fungal multi-domain GH3 beta-N-acetylglucosaminidase [Rhizomucor miehei CAU432] |
| 7VI6_A | 1.74e-40 | 67 | 341 | 5 | 279 | ChainA, Beta-N-acetylhexosaminidase [Akkermansia muciniphila ATCC BAA-835],7VI6_B Chain B, Beta-N-acetylhexosaminidase [Akkermansia muciniphila ATCC BAA-835],7VI7_A Chain A, Beta-N-acetylhexosaminidase [Akkermansia muciniphila ATCC BAA-835],7VI7_B Chain B, Beta-N-acetylhexosaminidase [Akkermansia muciniphila ATCC BAA-835] |
| 3BMX_A | 2.33e-38 | 50 | 378 | 36 | 394 | Beta-N-hexosaminidase(YbbD) from Bacillus subtilis [Bacillus subtilis],3BMX_B Beta-N-hexosaminidase (YbbD) from Bacillus subtilis [Bacillus subtilis],3NVD_A Structure of YBBD in complex with pugnac [Bacillus subtilis],3NVD_B Structure of YBBD in complex with pugnac [Bacillus subtilis] |
| 3TEV_A | 3.98e-38 | 58 | 372 | 14 | 330 | Thecrystal structure of glycosyl hydrolase from Deinococcus radiodurans R1 [Deinococcus radiodurans R1],3TEV_B The crystal structure of glycosyl hydrolase from Deinococcus radiodurans R1 [Deinococcus radiodurans R1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P48823 | 2.11e-40 | 74 | 377 | 50 | 380 | Beta-hexosaminidase A OS=Pseudoalteromonas piscicida OX=43662 GN=cht60 PE=1 SV=1 |
| A6VKU4 | 3.53e-40 | 66 | 352 | 4 | 293 | Beta-hexosaminidase OS=Actinobacillus succinogenes (strain ATCC 55618 / DSM 22257 / CCUG 43843 / 130Z) OX=339671 GN=nagZ PE=3 SV=1 |
| A3N1B7 | 4.81e-40 | 65 | 352 | 1 | 291 | Beta-hexosaminidase OS=Actinobacillus pleuropneumoniae serotype 5b (strain L20) OX=416269 GN=nagZ PE=3 SV=1 |
| B3GXZ7 | 4.81e-40 | 65 | 352 | 1 | 291 | Beta-hexosaminidase OS=Actinobacillus pleuropneumoniae serotype 7 (strain AP76) OX=537457 GN=nagZ PE=3 SV=1 |
| B0BQ51 | 6.72e-40 | 65 | 352 | 1 | 291 | Beta-hexosaminidase OS=Actinobacillus pleuropneumoniae serotype 3 (strain JL03) OX=434271 GN=nagZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
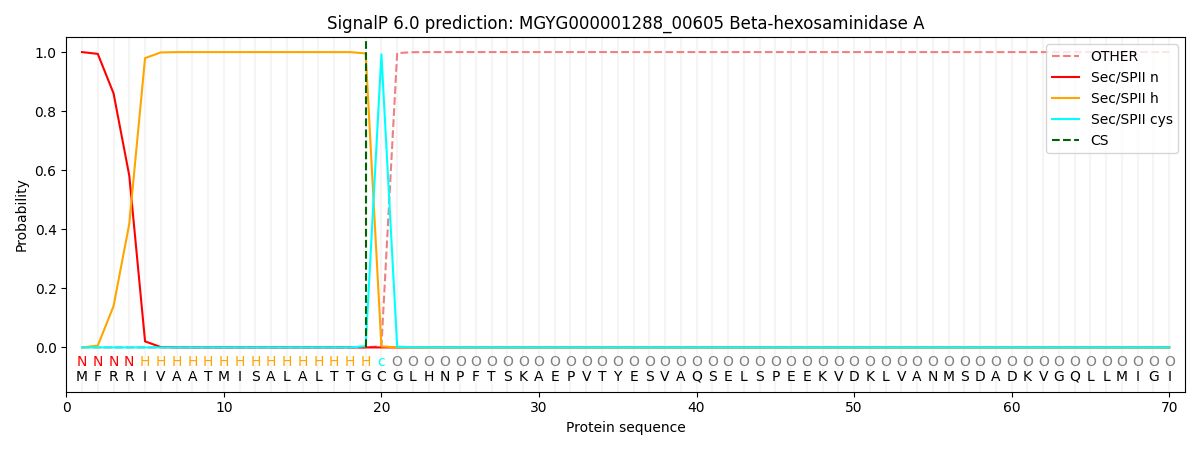
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000043 | 0.000000 | 0.000000 | 0.000000 |
