You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001378_00126
You are here: Home > Sequence: MGYG000001378_00126
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides ovatus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides ovatus | |||||||||||
| CAZyme ID | MGYG000001378_00126 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Beta-hexosaminidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 168166; End: 170619 Strand: - | |||||||||||
Full Sequence Download help
| MKKKIGGIGL CMLAWSILTC AQTITPQAEQ RAREIVSKMT LQEKIEYISG YTSFSLRAIP | 60 |
| RLGIPEIKLA DGPQGIRNHA PKSTLYPSGI LSASTWNRSL LYQLGQGLGQ DAKARGVNIL | 120 |
| LGPGVNIYRA PMCGRNFEYF GEDPYLTGET AKQYILGVQS EGVIATIKHF AANNQEWSRH | 180 |
| HASSDIDERT LHEIYFPAFR KAVQEANVSA VMNSYNLLNG VHATEHKWLN IDILRNLWGF | 240 |
| KGILMSDWTS VYSAVGAANA GLDLEMPKGR FMNVDNLIPA IKNGTVTEET INLKVQHILQ | 300 |
| TLIAYGMLDK EQKDSNIAQD NPFSRQAALE LAREGVVLLK NEGNLLPLKG KTAVMGPNAD | 360 |
| RIPTGGGSGF VTPFSTVSVS EGLEKLKKKN LVLLTDDVIY EDILHEFYAD AARQTKGFKA | 420 |
| EYFKNKMLSG QPEVTRTEAS VDYDWQYGAP LEGFPEDGFS VRWTASYMSQ KDGLLKLSIG | 480 |
| GDDGYRLFVN DKHITGDWGN HSYSSREVEL PVEAGKEYSF RIEFFDNISS AIIRFKASRL | 540 |
| NEEKLRQGLA KVDNVVFCTG FNSNTEGEGF DRPFALLHYQ ELFIQKVASL HPNLVVVLNA | 600 |
| GGGVDFTSWH EAAKAILMAW YPGQEGGQAI AEILTGKISP SGKLPISIEK KWEDNPVHDS | 660 |
| YYENLKAEIK RVDYSEGVFV GYRGYDRSGK EPFYPFGYGL SYSTFAYSNL AVEKTGEHQV | 720 |
| TVSFDIKNTG KMDASEIAQV YVHDVESSVP RPLKELKGYD KVFLKKGETQ RLSIVLNEDA | 780 |
| FSYYDMNQHR FVVEKGVFEI LVGPASNQLP LKAAIEL | 817 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 58 | 267 | 1e-68 | 0.9861111111111112 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK15098 | PRK15098 | 6.45e-80 | 61 | 803 | 101 | 751 | beta-glucosidase BglX. |
| COG1472 | BglX | 1.42e-78 | 57 | 368 | 53 | 372 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| PLN03080 | PLN03080 | 2.43e-57 | 30 | 777 | 51 | 743 | Probable beta-xylosidase; Provisional |
| pfam00933 | Glyco_hydro_3 | 2.16e-47 | 61 | 299 | 63 | 314 | Glycosyl hydrolase family 3 N terminal domain. |
| pfam01915 | Glyco_hydro_3_C | 9.36e-41 | 336 | 682 | 1 | 216 | Glycosyl hydrolase family 3 C-terminal domain. This domain is involved in catalysis and may be involved in binding beta-glucan. This domain is found associated with pfam00933. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGT72363.1 | 0.0 | 1 | 817 | 1 | 817 |
| ALJ47550.1 | 0.0 | 1 | 817 | 1 | 817 |
| QRQ54466.1 | 0.0 | 1 | 817 | 1 | 817 |
| QUT26117.1 | 0.0 | 1 | 817 | 1 | 817 |
| QDH53357.1 | 0.0 | 1 | 817 | 1 | 817 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7MS2_A | 9.36e-137 | 33 | 816 | 6 | 667 | ChainA, Thermostable beta-glucosidase B [Acetivibrio thermocellus AD2],7MS2_B Chain B, Thermostable beta-glucosidase B [Acetivibrio thermocellus AD2] |
| 3AC0_A | 1.69e-128 | 34 | 809 | 8 | 828 | Crystalstructure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus],3AC0_B Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus],3AC0_C Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus],3AC0_D Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus] |
| 3ABZ_A | 4.00e-125 | 34 | 809 | 8 | 828 | Crystalstructure of Se-Met labeled Beta-glucosidase from Kluyveromyces marxianus [Kluyveromyces marxianus],3ABZ_B Crystal structure of Se-Met labeled Beta-glucosidase from Kluyveromyces marxianus [Kluyveromyces marxianus],3ABZ_C Crystal structure of Se-Met labeled Beta-glucosidase from Kluyveromyces marxianus [Kluyveromyces marxianus],3ABZ_D Crystal structure of Se-Met labeled Beta-glucosidase from Kluyveromyces marxianus [Kluyveromyces marxianus] |
| 5WAB_A | 8.90e-117 | 36 | 817 | 9 | 663 | CrystalStructure of Bifidobacterium adolescentis GH3 beta-glucosidase [Bifidobacterium adolescentis ATCC 15703],5WAB_B Crystal Structure of Bifidobacterium adolescentis GH3 beta-glucosidase [Bifidobacterium adolescentis ATCC 15703],5WAB_C Crystal Structure of Bifidobacterium adolescentis GH3 beta-glucosidase [Bifidobacterium adolescentis ATCC 15703],5WAB_D Crystal Structure of Bifidobacterium adolescentis GH3 beta-glucosidase [Bifidobacterium adolescentis ATCC 15703] |
| 2X40_A | 1.78e-114 | 30 | 816 | 2 | 712 | Structureof beta-glucosidase 3B from Thermotoga neapolitana in complex with glycerol [Thermotoga neapolitana DSM 4359],2X41_A Structure of beta-glucosidase 3B from Thermotoga neapolitana in complex with glucose [Thermotoga neapolitana DSM 4359] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P27034 | 2.31e-146 | 34 | 817 | 4 | 808 | Beta-glucosidase OS=Rhizobium radiobacter OX=358 GN=cbg-1 PE=3 SV=1 |
| P14002 | 5.13e-136 | 33 | 816 | 6 | 667 | Thermostable beta-glucosidase B OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=bglB PE=1 SV=2 |
| Q5BFG8 | 8.68e-134 | 35 | 817 | 14 | 838 | Beta-glucosidase B OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=bglB PE=1 SV=1 |
| A1DNN8 | 2.58e-129 | 34 | 809 | 16 | 857 | Probable beta-glucosidase J OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=bglJ PE=3 SV=1 |
| B0Y8M8 | 1.02e-128 | 34 | 798 | 16 | 837 | Probable beta-glucosidase J OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=bglJ PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
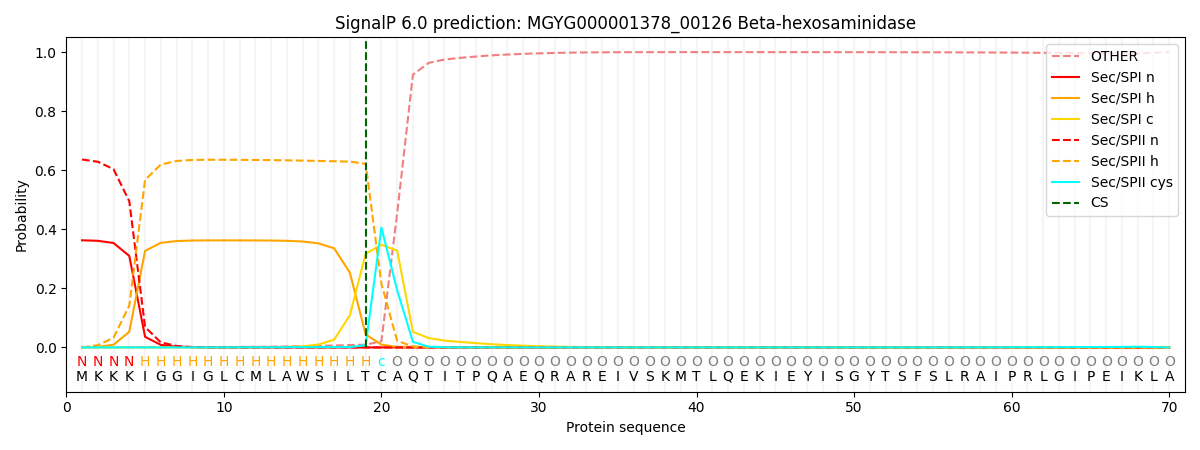
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.003825 | 0.349890 | 0.645055 | 0.000478 | 0.000365 | 0.000368 |
