You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001420_00064
You are here: Home > Sequence: MGYG000001420_00064
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes senegalensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; Alistipes senegalensis | |||||||||||
| CAZyme ID | MGYG000001420_00064 | |||||||||||
| CAZy Family | GH89 | |||||||||||
| CAZyme Description | Beta-hexosaminidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 79126; End: 83820 Strand: + | |||||||||||
Full Sequence Download help
| MSLRLTSLCR AVLLTLFLSL PALAGASPER TDFSGVARLV ERRIPFLAGK VDFRPASDAA | 60 |
| ENSDAFTLAT RRGRLTIEAT SPSAAATAVN YYLNRFCGMS LSHNGDNLHD LPSLPEVAPS | 120 |
| VTVRSPFRYR YALNYCTYNY TYSFYGWNDW ERELDWMALN GVNLMLAPLG TELVWARTLE | 180 |
| DAGFTEKEIG DFIPGPAFTA WWLMGNLEGW GGPMSRGMME SRVRMQQRIL ARMAELGIEP | 240 |
| VLQGFWGMVP NKLREKYPDA RIVDQGLWGR IFPRPAILLP EDPLFDRMSD RYYSVLRELY | 300 |
| GDSVKYLGGD LFHEGGDTTG MHVTRTASLI QASMQRHFPG SKWMLQGWSG NPKKELLAGL | 360 |
| DPALTVVIDL FGESGSTWRN TGEFYGTPWI WATVNHFGGK TDMGGQLPVI LAGPHEARSE | 420 |
| SENLLCGVGI LPEGIAANPV VYDWALKTAW EPGAPDPDDY LRRYIVYRYG TWNDHAYEAW | 480 |
| RLLLGSVYGE FAIKGEGTFE SIFCARPGLD VTSVSTWGPK RFQYDPAKLV DALVLFRKAA | 540 |
| EQYGDSPTYR YDLADLARQV LANHARDIYQ RSVKAYRRGD REGFGLLSAD FLALLGLQDR | 600 |
| LTGTQPDFLL GRWLDKALHY GDNEQERRQS LKNAKTQITY WGPADPNTRV HDYANKEWSG | 660 |
| LLADYYLPRW KAFFDYLSAD MEGRKVPEPD YFGMEKAWAD SGKEYPTEPR GELLAEVDSV | 720 |
| LLAVCPPYKD RSLTPQERAE DLLGRMTLRE KIAQMRHIHF KHYDNEGEVD LEKLASSTGG | 780 |
| VGWGCVEAFP YSSGQYLKAM RQIQAYMCSD TRLGIPVIPV MEGLHGVVQD GSTIFPQAIA | 840 |
| QGATFNPRLV GGMAGHIADE MEAIGAKQVL APVLDIAREL RWGRVEETFG EDPHLISCMG | 900 |
| AAYMKAMHAK GMITTPKHFV AHGTPTAGLN LASVKGGKRE LMSLYVKPFR ELIAATKPRS | 960 |
| AMNCYSSYDD EAVTGSKSLM TGLLRDSLGF RGYVYSDWGS VRMLSHFHRA AAPGGAAARM | 1020 |
| AIEAGIDLEA GSEEYRHAEQ MVREGFLDEK YIDRAVGNIL FVKFASGLFD GEPADTLGWR | 1080 |
| RKIHAPEAVR LARQIADESA VLLENRNGVL PLSPGKLRSV AVIGPNADRV QFGDYSWSAE | 1140 |
| KDKGITPLCG IREYLDGSGV RVNYAEGCDP YSQDKSGFGE ALKAARRSDA VVVFVGSQSA | 1200 |
| ILARASEPAT SGEGYDLTSL RLPGVQEELV EELARTGKPM IVVLVTGKPF ELARIRELAD | 1260 |
| AVLVQWYAGE EGGASVAGIL FGKTNPSGRL PVSFPQSAGH LPCYYNHLPT DKGYYNKKGT | 1320 |
| PDNPGRDYVF SDPEPLYPFG YGLSYTQFEY SGMETASGEF TATDTLRVNV RVKNTGSRTG | 1380 |
| SEVVQLYVRD LISSVATPVR QLHAFDKVEL NPGEERSVRF SIPVPLLSLY DAAMRRIVEP | 1440 |
| GDFELQVGAS SRDIRLRDTV TVAGCADIPG PKAIGAKAGA VREPGAEVEI SGTVRNVQAA | 1500 |
| VMPDVRVYAA SAPACAVVTD RHGRYTLRIR TNDRAVFELE GHERREIDVT EGGRLDIELE | 1560 |
| PAMN | 1564 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH89 | 72 | 720 | 3e-207 | 0.9924585218702866 |
| GH3 | 810 | 1029 | 2.1e-60 | 0.9722222222222222 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam05089 | NAGLU | 2.65e-146 | 130 | 452 | 1 | 333 | Alpha-N-acetylglucosaminidase (NAGLU) tim-barrel domain. Alpha-N-acetylglucosaminidase, a lysosomal enzyme required for the stepwise degradation of heparan sulfate. Mutations on the alpha-N-acetylglucosaminidase (NAGLU) gene can lead to Mucopolysaccharidosis type IIIB (MPS IIIB; or Sanfilippo syndrome type B) characterized by neurological dysfunction but relatively mild somatic manifestations. The structure shows that the enzyme is composed of three domains. This central domain has a tim barrel fold. |
| PRK15098 | PRK15098 | 1.25e-144 | 733 | 1455 | 29 | 758 | beta-glucosidase BglX. |
| pfam12972 | NAGLU_C | 1.13e-95 | 460 | 717 | 1 | 257 | Alpha-N-acetylglucosaminidase (NAGLU) C-terminal domain. Alpha-N-acetylglucosaminidase, a lysosomal enzyme required for the stepwise degradation of heparan sulfate. Mutations on the alpha-N-acetylglucosaminidase (NAGLU) gene can lead to Mucopolysaccharidosis type IIIB (MPS IIIB; or Sanfilippo syndrome type B) characterized by neurological dysfunction but relatively mild somatic manifestations. The structure shows that the enzyme is composed of three domains. This C-terminal domain has an all alpha helical fold. |
| PLN03080 | PLN03080 | 1.28e-81 | 727 | 1448 | 41 | 769 | Probable beta-xylosidase; Provisional |
| COG1472 | BglX | 4.22e-76 | 746 | 1145 | 1 | 379 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBL07501.1 | 0.0 | 3 | 1564 | 2 | 1563 |
| ALO49402.1 | 0.0 | 25 | 1559 | 25 | 1557 |
| QUB47602.1 | 0.0 | 16 | 1559 | 10 | 1549 |
| QIL38350.1 | 3.97e-297 | 726 | 1562 | 25 | 862 |
| QNL52448.1 | 7.17e-255 | 726 | 1549 | 25 | 849 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5YOT_A | 4.62e-146 | 726 | 1455 | 5 | 749 | Isoprimeverose-producingenzyme from Aspergillus oryzae in complex with isoprimeverose [Aspergillus oryzae RIB40],5YOT_B Isoprimeverose-producing enzyme from Aspergillus oryzae in complex with isoprimeverose [Aspergillus oryzae RIB40],5YQS_A Isoprimeverose-producing enzyme from Aspergillus oryzae in complex with isoprimeverose [Aspergillus oryzae RIB40],5YQS_B Isoprimeverose-producing enzyme from Aspergillus oryzae in complex with isoprimeverose [Aspergillus oryzae RIB40] |
| 7EAP_A | 3.32e-145 | 726 | 1455 | 5 | 749 | ChainA, Fn3_like domain-containing protein [Aspergillus oryzae RIB40] |
| 2VC9_A | 1.89e-137 | 73 | 722 | 212 | 874 | Family89 Glycoside Hydrolase from Clostridium perfringens in complex with 2-acetamido-1,2-dideoxynojirmycin [Clostridium perfringens],2VCA_A Family 89 glycoside hydrolase from Clostridium perfringens in complex with beta-N-acetyl-D-glucosamine [Clostridium perfringens],2VCB_A Family 89 Glycoside Hydrolase from Clostridium perfringens in complex with PUGNAc [Clostridium perfringens],2VCC_A Family 89 Glycoside Hydrolase from Clostridium perfringens [Clostridium perfringens] |
| 7MFK_A | 2.32e-137 | 73 | 722 | 220 | 882 | ChainA, Alpha-N-acetylglucosaminidase family protein [Clostridium perfringens ATCC 13124],7MFL_A Chain A, Alpha-N-acetylglucosaminidase family protein [Clostridium perfringens ATCC 13124] |
| 4A4A_A | 2.40e-136 | 73 | 722 | 235 | 897 | CpGH89(E483Q, E601Q), from Clostridium perfringens, in complex with its substrate GlcNAc-alpha-1,4-galactose [Clostridium perfringens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KMH0 | 9.34e-118 | 809 | 1452 | 59 | 711 | Beta-xylosidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22130 PE=1 SV=1 |
| Q9FNA3 | 1.83e-110 | 34 | 715 | 47 | 795 | Alpha-N-acetylglucosaminidase OS=Arabidopsis thaliana OX=3702 GN=NAGLU PE=2 SV=1 |
| Q56078 | 5.53e-109 | 728 | 1455 | 24 | 758 | Periplasmic beta-glucosidase OS=Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) OX=99287 GN=bglX PE=3 SV=2 |
| T2KMH9 | 1.56e-108 | 728 | 1455 | 29 | 748 | Putative beta-xylosidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22230 PE=2 SV=1 |
| P54802 | 7.65e-105 | 25 | 728 | 21 | 738 | Alpha-N-acetylglucosaminidase OS=Homo sapiens OX=9606 GN=NAGLU PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
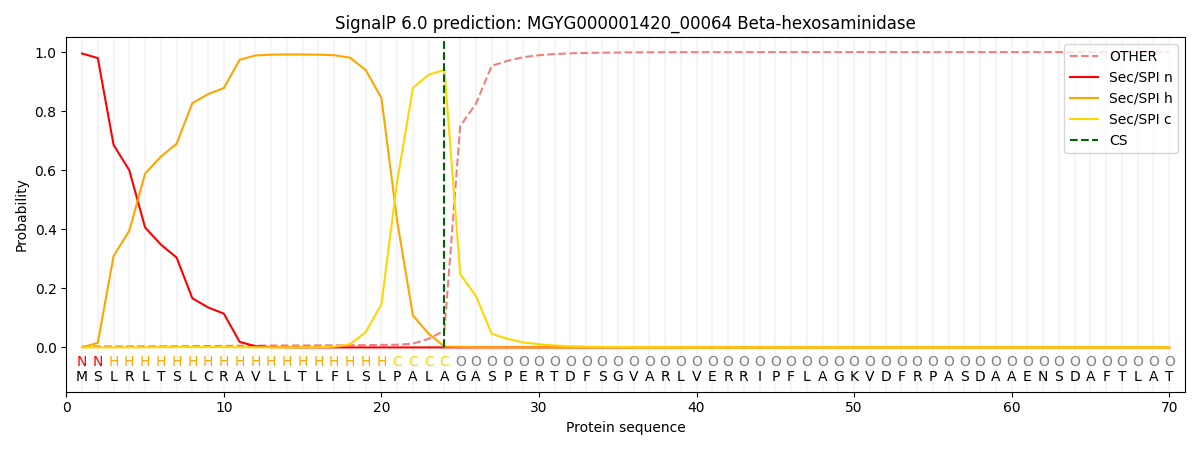
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.004478 | 0.993447 | 0.001445 | 0.000214 | 0.000197 | 0.000182 |
