You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001527_00139
You are here: Home > Sequence: MGYG000001527_00139
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
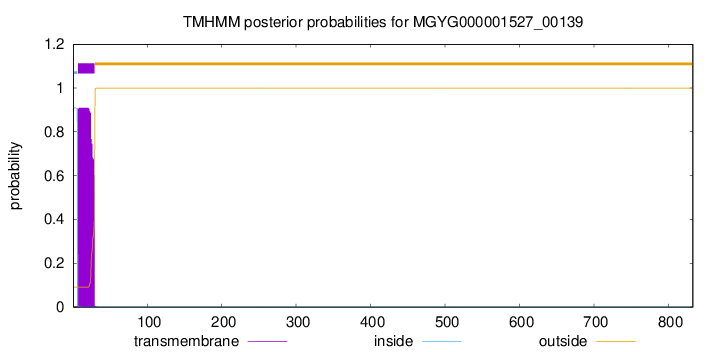
TMHMM annotations
Basic Information help
| Species | Fournierella massiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Fournierella; Fournierella massiliensis | |||||||||||
| CAZyme ID | MGYG000001527_00139 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 21782; End: 24283 Strand: - | |||||||||||
Full Sequence Download help
| MKNVQRFLAF VLAIGMALSM AACGQTPAST PASSSTSTAA EPSGYMPEPS EGAKYTVTTT | 60 |
| EDGWLKLENE GGEVLGLSPN SGVKVIEDDG YAFKDLNQNG TLDAYEDWRL TTEERTADLV | 120 |
| GQMEGAEMAA ILSHGGWGDF TTEPLAEDDG SRTFLEAGGR GGVTRNIGRG GGVHAQWTNQ | 180 |
| IQAVAESCYY GIPAMISIDP ANISGLVETT ALASTMDPEL AAEIGVETSK EYRAAGVTAL | 240 |
| LGPQVDIAGA SMSRAGGTYG EDPQLTLDIA TAYVNAMQST YDENGEDLGW GDESVFCFTK | 300 |
| HYAGAGATEG GRDDHHYTAH YAVFPGDNLE AHLITYFDGV FNLPGKTGSS GIMTQYSVNV | 360 |
| DGEGNPYGGE DLAGAYNAYM YGLLDEAGYD SLTITDWGVF QFAGCWGTED MADEAEKIAF | 420 |
| GWERGVELLG GYGTMETVAA AYDKLVERNG EETAKEIISH AASNYIEVMM NLNMFDQPYN | 480 |
| DSAYAESIIT TGGNSELGLE GQRDSVVMLK NDGTIKEGGS ATSDKPTVYI PYVYNTGFSV | 540 |
| SWMGGVSEGT PSWTPGMDLD VMGQYFNVVT DKLGDPTGEA DAEGNPTYTK DDLTRATAEE | 600 |
| IAACDYILVG MSNPYSISYT DPYVSAFGYL EAYKNNELYP DDIYWYPVSM QYGEYTAETA | 660 |
| RETSIAGPIV DGEKQNRSYK GNTAPAAANY GDLEALQYAD SVAGDIPVIV SMKMERGMVW | 720 |
| TETESMADVI LGCYSKQRPE VVAEIILGET EPNGLLVFQQ PADMEAVEAQ LEDVPRDMEC | 780 |
| YVDANGNTYD FAFGLNWAGV INDERVEKYS AEPLTKVQNI DYAAYAASYG AEG | 833 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 191 | 400 | 1.7e-25 | 0.8611111111111112 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK15098 | PRK15098 | 4.55e-12 | 211 | 513 | 125 | 402 | beta-glucosidase BglX. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCJ97597.1 | 2.27e-209 | 54 | 823 | 82 | 848 |
| QGY45729.1 | 4.14e-142 | 38 | 814 | 33 | 832 |
| QIA06569.1 | 9.78e-142 | 47 | 814 | 42 | 827 |
| AQR97785.1 | 2.94e-139 | 46 | 809 | 25 | 799 |
| AGF59116.1 | 4.13e-139 | 46 | 809 | 25 | 799 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q46684 | 3.96e-40 | 82 | 515 | 35 | 490 | Periplasmic beta-glucosidase/beta-xylosidase OS=Dickeya chrysanthemi OX=556 GN=bgxA PE=3 SV=1 |
| Q5BCC6 | 2.00e-32 | 95 | 779 | 28 | 603 | Beta-glucosidase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=bglC PE=1 SV=1 |
| Q2UFP8 | 5.44e-31 | 90 | 798 | 33 | 627 | Probable beta-glucosidase C OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=bglC PE=3 SV=2 |
| B8NGU6 | 2.95e-30 | 105 | 798 | 42 | 623 | Probable beta-glucosidase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=bglC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
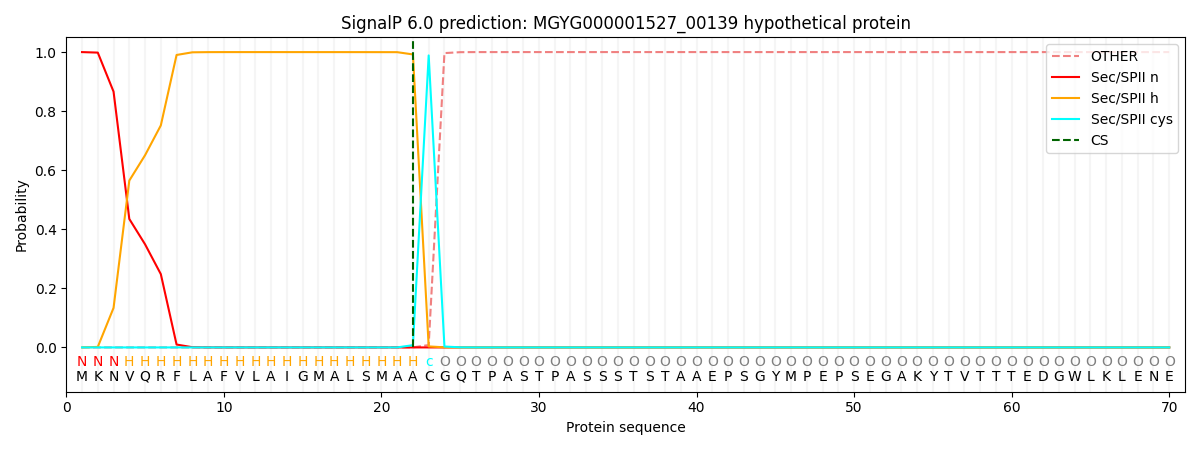
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000002 | 1.000049 | 0.000000 | 0.000000 | 0.000000 |