You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001663_00019
You are here: Home > Sequence: MGYG000001663_00019
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes sp900760675 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; Alistipes sp900760675 | |||||||||||
| CAZyme ID | MGYG000001663_00019 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Beta-glucosidase BoGH3A | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 18890; End: 21094 Strand: - | |||||||||||
Full Sequence Download help
| MGACMFAGSV VAQALPAYMD DKKPIEQRIE DAIGRMTLKE KVAMLHAQSK FSSPGVAKLG | 60 |
| IPEFWMTDGP HGIRPEVMWD EWEQAGWTND SCVAFPALTC LAASWNPEMS LLYGRSIGEE | 120 |
| ARYRNKAVLL GPGVNIYRTP LNGRNFEYMG EDPYLASRMV VPYIQGVQQN GVAACVKHYA | 180 |
| LNNQEVNRHT TNVIVGDRAL YEIYLPAFRA AVQEGGAWSI MGSYNLYKEE HGCHNQYLLN | 240 |
| DILKDEWKFD GVVVSDWGGT HDTWQAIHNG LDMEFGSWTD GLSNGASNAY DNYYLADPYL | 300 |
| RLIAEGKVGT NELDDKVRRI LRLAFRTTMD RNRPFGSMCS DEHFDAARRI GQEGIVLLQN | 360 |
| NGNVLPIDTE CARKILVVGE NAVKMMTVGG GSSSLKVKYE ISPLDGIRKR FGDKAEVTYV | 420 |
| RGYVGDVTGE YNGVTSGQDL KDDRTPEELI GEAVGAARTA DYVIFVGGLN KSAHQDCEDA | 480 |
| DRLGLELPYD QDKVIAALAK ANKNLIVVNI SGNAVAMPWR AEVPAIVQAW YLGSETGNAL | 540 |
| AAILSGDANP SGKLPFTFYA SLDQCGAHAA GEYPGVPRGD GVVDQKYNEG IFVGYRWVDK | 600 |
| QNIKPLFPFG HGLSYTTFKY GKAVADKAVI TADQTLTVTI PVTNTGAREG AEVVQCYIRD | 660 |
| LKSSLVRPVK ELKGFGKIYL KPGETGNVTF TIGNDALCFF DDSRHEWVSE PGKFEVLVGA | 720 |
| SAQDIRAKVG FELK | 734 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 56 | 274 | 9e-66 | 0.9722222222222222 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK15098 | PRK15098 | 6.02e-118 | 24 | 733 | 30 | 764 | beta-glucosidase BglX. |
| pfam01915 | Glyco_hydro_3_C | 1.02e-69 | 355 | 595 | 1 | 216 | Glycosyl hydrolase family 3 C-terminal domain. This domain is involved in catalysis and may be involved in binding beta-glucan. This domain is found associated with pfam00933. |
| COG1472 | BglX | 3.78e-68 | 39 | 413 | 43 | 389 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| PLN03080 | PLN03080 | 7.36e-64 | 24 | 692 | 48 | 742 | Probable beta-xylosidase; Provisional |
| pfam00933 | Glyco_hydro_3 | 7.97e-33 | 40 | 323 | 46 | 316 | Glycosyl hydrolase family 3 N terminal domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT74223.1 | 0.0 | 1 | 734 | 8 | 741 |
| QCQ43197.1 | 0.0 | 1 | 734 | 11 | 744 |
| AKA53261.1 | 0.0 | 1 | 734 | 11 | 744 |
| CUA20132.1 | 0.0 | 1 | 734 | 11 | 744 |
| QUT89173.1 | 0.0 | 12 | 734 | 18 | 740 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7MS2_A | 3.07e-149 | 29 | 728 | 5 | 663 | ChainA, Thermostable beta-glucosidase B [Acetivibrio thermocellus AD2],7MS2_B Chain B, Thermostable beta-glucosidase B [Acetivibrio thermocellus AD2] |
| 5WAB_A | 5.32e-122 | 32 | 727 | 8 | 657 | CrystalStructure of Bifidobacterium adolescentis GH3 beta-glucosidase [Bifidobacterium adolescentis ATCC 15703],5WAB_B Crystal Structure of Bifidobacterium adolescentis GH3 beta-glucosidase [Bifidobacterium adolescentis ATCC 15703],5WAB_C Crystal Structure of Bifidobacterium adolescentis GH3 beta-glucosidase [Bifidobacterium adolescentis ATCC 15703],5WAB_D Crystal Structure of Bifidobacterium adolescentis GH3 beta-glucosidase [Bifidobacterium adolescentis ATCC 15703] |
| 4I3G_A | 7.14e-110 | 5 | 726 | 29 | 821 | CrystalStructure of DesR, a beta-glucosidase from Streptomyces venezuelae in complex with D-glucose. [Streptomyces venezuelae],4I3G_B Crystal Structure of DesR, a beta-glucosidase from Streptomyces venezuelae in complex with D-glucose. [Streptomyces venezuelae] |
| 2X40_A | 1.24e-107 | 27 | 733 | 2 | 713 | Structureof beta-glucosidase 3B from Thermotoga neapolitana in complex with glycerol [Thermotoga neapolitana DSM 4359],2X41_A Structure of beta-glucosidase 3B from Thermotoga neapolitana in complex with glucose [Thermotoga neapolitana DSM 4359] |
| 3AC0_A | 8.36e-107 | 29 | 734 | 6 | 837 | Crystalstructure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus],3AC0_B Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus],3AC0_C Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus],3AC0_D Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A7LXS8 | 2.64e-286 | 16 | 732 | 33 | 744 | Beta-glucosidase BoGH3A OS=Bacteroides ovatus (strain ATCC 8483 / DSM 1896 / JCM 5824 / BCRC 10623 / CCUG 4943 / NCTC 11153) OX=411476 GN=BACOVA_02644 PE=1 SV=1 |
| P14002 | 1.68e-148 | 29 | 728 | 5 | 663 | Thermostable beta-glucosidase B OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=bglB PE=1 SV=2 |
| P27034 | 5.98e-130 | 29 | 733 | 2 | 810 | Beta-glucosidase OS=Rhizobium radiobacter OX=358 GN=cbg-1 PE=3 SV=1 |
| Q5BFG8 | 1.94e-122 | 19 | 734 | 1 | 839 | Beta-glucosidase B OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=bglB PE=1 SV=1 |
| F6C6C1 | 1.42e-119 | 32 | 734 | 8 | 673 | Exo-alpha-(1->6)-L-arabinopyranosidase OS=Bifidobacterium breve (strain ACS-071-V-Sch8b) OX=866777 GN=HMPREF9228_1477 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
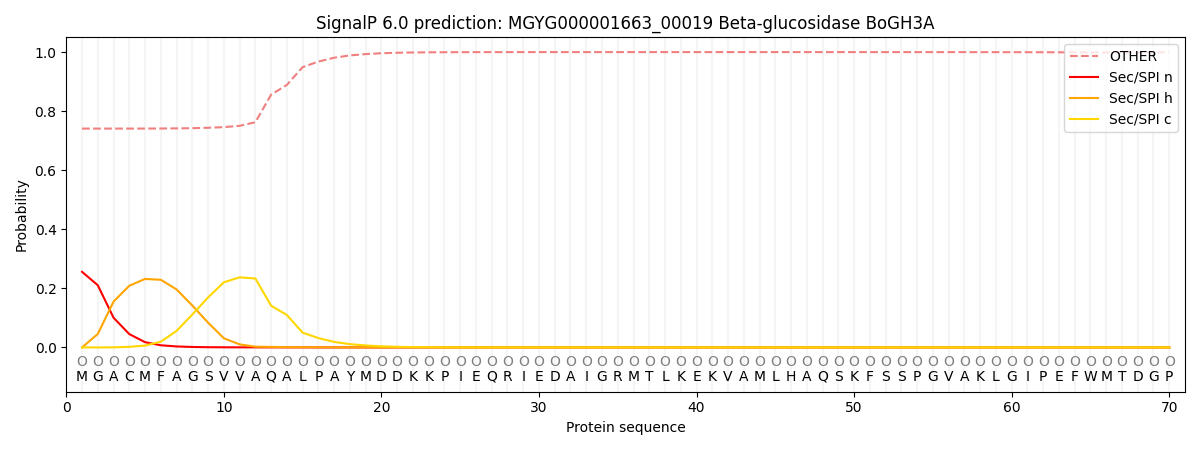
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.752192 | 0.242702 | 0.004024 | 0.000466 | 0.000273 | 0.000352 |
