You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001698_00321
You are here: Home > Sequence: MGYG000001698_00321
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
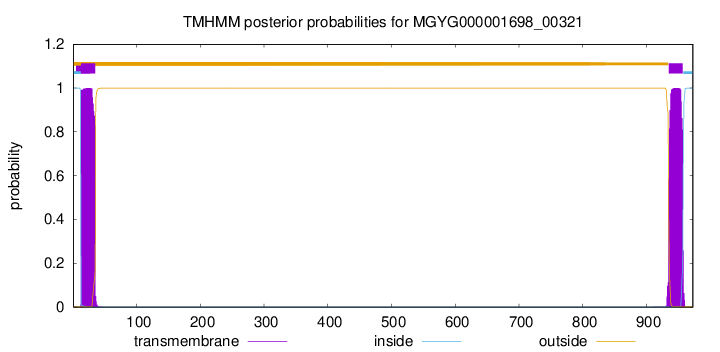
TMHMM annotations
Basic Information help
| Species | Marvinbryantia formatexigens | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Marvinbryantia; Marvinbryantia formatexigens | |||||||||||
| CAZyme ID | MGYG000001698_00321 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 328949; End: 331870 Strand: - | |||||||||||
Full Sequence Download help
| MSAKTKKMKK SKAVLAWGIS AVLIGSALGA GNYLAYHSFS GLADTVFGGV RPVYNTDVIS | 60 |
| MYPATNASSK EEAFQNAQEV NLRVAEEGFV LLKNEAEALP LQKGAKISVF GKNSVNLSYG | 120 |
| GSGSGGFDTS NNQNLYDSLE NAGFDINQTL KKFYEDNGAS GPARSANSSD LDSGDNQRIA | 180 |
| TAETPQSKYT DAVKASYDSY KDAALVVITR IGGEGFDLPR YQGTTEGAVS ADSHYLELDQ | 240 |
| NERDLLAAVC EEGFEHVVVV FNVPSSFEAT FLKDSAYAAF SDKVDAAVWI GFTGSNGITA | 300 |
| LGEILNGEVN PSGRLVDTWA SDFTQNPSFQ NFGTGHTDQT TDKYDAGMYY SVDYEEGIYV | 360 |
| GYRYYETRGL TDGEEWYEEN VTYPFGYGLS YTTFEWNVGE ASADEFDLES VITVPVTVKN | 420 |
| TGDTAGKEVV QLYVSAPYTE GGIEKAHKVL AGYAKTSLLQ PGESETVTVS FEPYSAASYD | 480 |
| YRDANENGFC GYELEAGEYT LYVSRNAHEE ESAITLTLLE DIQIGEDPVT GNEVVNRYTG | 540 |
| IEDNLDSDWQ LDTVLSRADW EGTWPTAQTD ETHAGSAELL AQIEDIETNN PTDFEEEDAP | 600 |
| WFGEDVISVL RDLLPETPAA DYHAIVDYDD DRWEEILDAC SEEEMLELIN NGAYHTKALE | 660 |
| TVGLPATIHG DGPAGFTCFM NSEQVNGTCQ YVSEPVMAAT WNDALLEEIG EAMGEEGILG | 720 |
| DKATSQPYSS IYAPGVNIHR SPFGGRCSEY YSEDPYISGM MASAQIRGLQ AKGVVPMLKH | 780 |
| FAANEQETHR SIGGDLSWVT EQSLREIYLK PFELAVKNGE SRGIMSSFNR IGARWTGGDY | 840 |
| RLLTEILRDE WGFRGTVICD FNTIPQYMVP RQMFYAGGDL DLATLETSMW TDADTSENGD | 900 |
| AIVLRNATKN ILYSFVNSNA MNAEVIGYNP PLWHGYLLWL DIGVAALLLL WGVLAFIHRA | 960 |
| KHNKKLQSMT ADS | 973 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 660 | 882 | 1.8e-50 | 0.9722222222222222 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1472 | BglX | 3.12e-34 | 697 | 860 | 89 | 248 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| PRK15098 | PRK15098 | 1.64e-28 | 65 | 468 | 376 | 722 | beta-glucosidase BglX. |
| pfam00933 | Glyco_hydro_3 | 7.23e-26 | 644 | 860 | 46 | 254 | Glycosyl hydrolase family 3 N terminal domain. |
| pfam01915 | Glyco_hydro_3_C | 8.63e-25 | 90 | 362 | 2 | 216 | Glycosyl hydrolase family 3 C-terminal domain. This domain is involved in catalysis and may be involved in binding beta-glucan. This domain is found associated with pfam00933. |
| PRK15098 | PRK15098 | 8.92e-17 | 697 | 864 | 126 | 291 | beta-glucosidase BglX. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| VEU80232.1 | 1.58e-269 | 44 | 965 | 46 | 962 |
| QOS39239.1 | 9.62e-261 | 10 | 961 | 5 | 964 |
| VEU80230.1 | 3.57e-199 | 65 | 921 | 31 | 910 |
| SQH20118.1 | 1.47e-188 | 54 | 971 | 59 | 971 |
| ASE08187.2 | 1.47e-188 | 54 | 971 | 59 | 971 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5WUG_A | 6.73e-83 | 85 | 929 | 44 | 793 | Expression,characterization and crystal structure of a novel beta-glucosidase from Paenibacillus barengoltzii [Paenibacillus barengoltzii],5WVP_A Expression, characterization and crystal structure of a novel beta-glucosidase from Paenibacillus barengoltzii [Paenibacillus barengoltzii] |
| 2X40_A | 2.10e-43 | 649 | 880 | 36 | 259 | Structureof beta-glucosidase 3B from Thermotoga neapolitana in complex with glycerol [Thermotoga neapolitana DSM 4359],2X41_A Structure of beta-glucosidase 3B from Thermotoga neapolitana in complex with glucose [Thermotoga neapolitana DSM 4359] |
| 2X42_A | 2.13e-42 | 649 | 880 | 36 | 259 | Structureof beta-glucosidase 3B from Thermotoga neapolitana in complex with alpha-D-glucose [Thermotoga neapolitana DSM 4359] |
| 5WAB_A | 6.04e-38 | 643 | 862 | 15 | 234 | CrystalStructure of Bifidobacterium adolescentis GH3 beta-glucosidase [Bifidobacterium adolescentis ATCC 15703],5WAB_B Crystal Structure of Bifidobacterium adolescentis GH3 beta-glucosidase [Bifidobacterium adolescentis ATCC 15703],5WAB_C Crystal Structure of Bifidobacterium adolescentis GH3 beta-glucosidase [Bifidobacterium adolescentis ATCC 15703],5WAB_D Crystal Structure of Bifidobacterium adolescentis GH3 beta-glucosidase [Bifidobacterium adolescentis ATCC 15703] |
| 7MS2_A | 2.47e-36 | 69 | 509 | 292 | 659 | ChainA, Thermostable beta-glucosidase B [Acetivibrio thermocellus AD2],7MS2_B Chain B, Thermostable beta-glucosidase B [Acetivibrio thermocellus AD2] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P16084 | 1.14e-87 | 63 | 861 | 14 | 770 | Beta-glucosidase A OS=Butyrivibrio fibrisolvens OX=831 GN=bglA PE=3 SV=1 |
| P15885 | 6.15e-82 | 72 | 861 | 8 | 697 | Beta-glucosidase OS=Ruminococcus albus OX=1264 PE=3 SV=1 |
| E7CY69 | 3.52e-38 | 643 | 862 | 15 | 234 | Exo-alpha-(1->6)-L-arabinopyranosidase OS=Bifidobacterium longum OX=216816 GN=apy PE=1 SV=1 |
| F6C6C1 | 8.27e-38 | 643 | 862 | 15 | 234 | Exo-alpha-(1->6)-L-arabinopyranosidase OS=Bifidobacterium breve (strain ACS-071-V-Sch8b) OX=866777 GN=HMPREF9228_1477 PE=1 SV=1 |
| P14002 | 1.36e-35 | 69 | 509 | 292 | 659 | Thermostable beta-glucosidase B OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=bglB PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
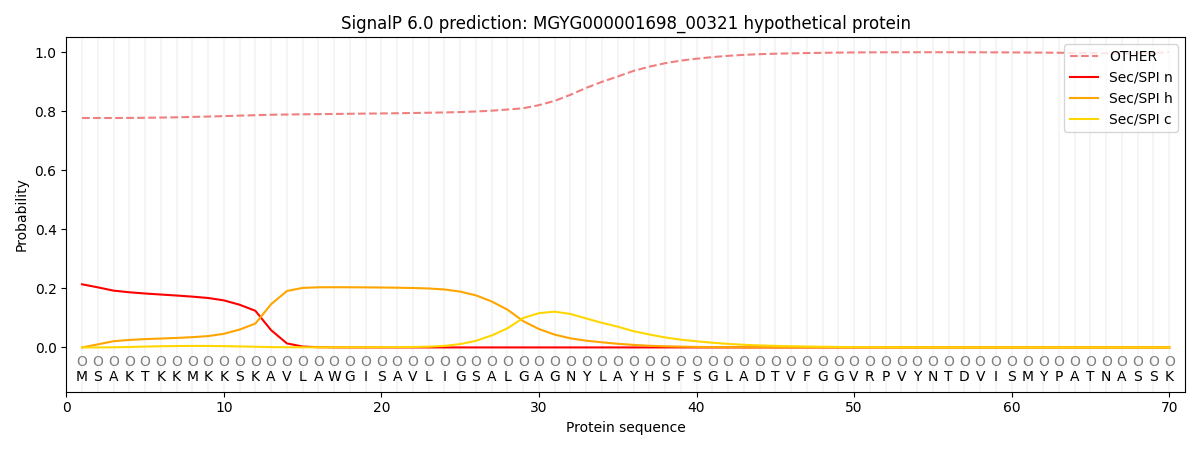
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.788311 | 0.198744 | 0.006173 | 0.000876 | 0.000432 | 0.005476 |