You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002135_00008
You are here: Home > Sequence: MGYG000002135_00008
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Anaeromassilibacillus sp002159845 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Acutalibacteraceae; Anaeromassilibacillus; Anaeromassilibacillus sp002159845 | |||||||||||
| CAZyme ID | MGYG000002135_00008 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Beta-hexosaminidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 8931; End: 15161 Strand: - | |||||||||||
Full Sequence Download help
| MKRWKKFISS ILAIAIATST LPASFVSFAQ DNSFKIEAET GYMVDRDGEK LDQAPATDDQ | 60 |
| HASGGAYYGH FDDGAGRGLI LENVSETDAT SLSIKYASYN QGTFDIYINA SEEAYQLAFI | 120 |
| ATGDWYMDTG REVSANIEIP AGATIKIVPS RPANIDYITL GSRAADSELI EAETGYMVDR | 180 |
| NGNQIDQAPA TDDQYASGGA YYGHFDEGEE KGLVLENVTD TAKTSLSIGY ASYNQGTFDI | 240 |
| YINDSESAYQ LSFTATGDWY MNTGREASGR IEIPAGATIR IVPSRPANID YIVLGTETDL | 300 |
| PAIPNEDDFV LIEAETGYMV DRGGNRIDQA PATDDEYASG GAYYDHFDSG ERKGLILENN | 360 |
| TGEDKTSIAI RYASKGYTGT FDLYINDSKT ARQLAFTSTS DWYMNTSNIV SGRIDIPDGA | 420 |
| TIKIVPSNPA NIDYIVLGND ETLPVIPEET VSREQELARQ METESIVLAQ NDGVLPLSED | 480 |
| TKIAVFGSGQ LAPATGGSGS GAVNGEYTSN FIDGLYELGI EPYQELLEYY EARVHSNNDI | 540 |
| DHGWDTSTEY PDEWGTPVYS GTSWSHAAGV NTPDVKLDNG EITDDGIVAR AAQESDVAIV | 600 |
| YITRTTGAEE MDRIEQPGDW NLNASERVLL EQVSDKFDRI VAILSINGPI EMSWVKEYGV | 660 |
| DAVMISYASG SQNGYAMADL VFGKENPSAK LADTIVETYD EHPTADTFGY ITYSEMGLDG | 720 |
| EANHTAFGDE DPISPYLENI YVGYRYFDTF GKEVLYPFGS GLSYSDFAFE DMDVGLNAAD | 780 |
| KSITVSATIE NVTEDPSIVP GKEVMEVYVS IPDGKLEQPY QKLVDYEKSE ELAAGESQRI | 840 |
| SIDVPLKDLA SYDEERAAYI LEPGTYYLRV GSSSRDTHIA GAFTIEDTIV VEQLTNQLVM | 900 |
| SDDAKALFEE KALTKEGATP ITYDGEAEEM RKAEAEAFTV TADDVEVSLE VPTVTEPSFR | 960 |
| ELPEDEPVHK FSEVQSGDIS LEDFVAQMTE DELITFLSGG NYRKDDGVGY SDDSGIPENF | 1020 |
| ASKASGVGGA GQTRNIERFS IPSITFADGS AGIGYSAIPG VRDKNIGWPR AAAMACIWNK | 1080 |
| ELLREFAVEM GKAMLDINVD VWLAPSINLH RNPLGGRGNE YYSEDPILSG LVASVVAEGV | 1140 |
| AESGVTVCLK HFAGNDQEYY RRGVINDTTE QNGTSRDAIN VIATERTLRE IYLKPFEMAV | 1200 |
| KTGKVMNVMS AFNKINGQYC ASNEGLLTDI LRGEWGFQGF VVTDWGDYDV LAHEGHALAA | 1260 |
| GNDLNMPGGH SRYWITNKYR ESLADGSLTL DQMRRSAYRL LNTIQSSALS SMPEKHNFAS | 1320 |
| NLSIQSTTLP DAKIGFEYSE SKVNPLIAAG GKGSRYSFQV ASDSPAQLPE GITLRPNGAL | 1380 |
| TGIPAAGTEG SYDIKFQVTD NTGASATKQL TLTVNGELSI APEELPVVVL NEAYEQKLTA | 1440 |
| SDAQGNPVDA IFTTTDALPT GLTLSEDGTI RGTCTDIGDG VFDITISVVS KDGSKTGTVS | 1500 |
| YKLDAISSTA QIATESLEDA VPGEFYSFAL SADGGVQPYT WEVSALPEGF VLHDGAIKSG | 1560 |
| RLSGKDFLEQ TVPESAEGVY EIEVKLTDSI GYEDTKTFLL KVGNPDEEAF AITTGSLGTG | 1620 |
| RVGASYETVL TSINGEGDVR YTLAEDSDSL PDGMTLSEDG ILSGTPSLLS SNLYRIVIHA | 1680 |
| TDGSGSTCDR TYSLYIAGQL TADPAAFQVL NASEGKAFKQ SFSATGGYNN EYKFELAEGS | 1740 |
| SALPAGLSMT SDSSGMTISG TPEPGTAGTY ELLILMDAAE FGGNPVTSVI PYTLVVEPEE | 1800 |
| INLALEKDVY ASSYIDMPPE WAVDGNMGTR WSCYWQDGDV DTPWLTVDLG ADYAIYRAVI | 1860 |
| YWETAQASEY VIQVSDDNET FTPVDVVGRS LNGNTHTWEI EATGRYIRMQ SIKAATKWGS | 1920 |
| SIFEFEVYGT PAEETDYSIA SKTYAIDAEN AIVSGIPAKT TAEDVLSELT CQAQDATIQI | 1980 |
| VDASGEALDG GGMVASQMKV QVVAHNEVRK EYVVSVLGDL DGSGGPSETA IEQIQSYILG | 2040 |
| DASLNLLKVL SADVKPDGKI NVQDLVLIKK SILLGQ | 2076 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 1035 | 1267 | 6.3e-61 | 0.9861111111111112 |
| CBM32 | 1816 | 1926 | 1.3e-23 | 0.8629032258064516 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1472 | BglX | 3.26e-44 | 1019 | 1267 | 39 | 279 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| pfam00933 | Glyco_hydro_3 | 1.18e-30 | 1069 | 1302 | 89 | 315 | Glycosyl hydrolase family 3 N terminal domain. |
| pfam01915 | Glyco_hydro_3_C | 1.88e-26 | 466 | 713 | 1 | 216 | Glycosyl hydrolase family 3 C-terminal domain. This domain is involved in catalysis and may be involved in binding beta-glucan. This domain is found associated with pfam00933. |
| PRK15098 | PRK15098 | 2.21e-23 | 458 | 885 | 387 | 764 | beta-glucosidase BglX. |
| pfam14310 | Fn3-like | 2.56e-20 | 803 | 874 | 1 | 71 | Fibronectin type III-like domain. This domain has a fibronectin type III-like structure. It is often found in association with pfam00933 and pfam01915. Its function is unknown. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCJ99238.1 | 4.53e-148 | 457 | 1307 | 13 | 854 |
| QRT50981.1 | 6.81e-144 | 455 | 1305 | 4 | 817 |
| VCV22851.1 | 9.82e-144 | 455 | 1305 | 4 | 830 |
| CBL11433.1 | 9.82e-144 | 455 | 1305 | 4 | 830 |
| CBL13855.1 | 8.78e-143 | 457 | 1305 | 6 | 817 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5WUG_A | 8.91e-109 | 466 | 1300 | 49 | 815 | Expression,characterization and crystal structure of a novel beta-glucosidase from Paenibacillus barengoltzii [Paenibacillus barengoltzii],5WVP_A Expression, characterization and crystal structure of a novel beta-glucosidase from Paenibacillus barengoltzii [Paenibacillus barengoltzii] |
| 2X42_A | 9.40e-58 | 451 | 885 | 321 | 713 | Structureof beta-glucosidase 3B from Thermotoga neapolitana in complex with alpha-D-glucose [Thermotoga neapolitana DSM 4359] |
| 2X40_A | 9.40e-58 | 451 | 885 | 321 | 713 | Structureof beta-glucosidase 3B from Thermotoga neapolitana in complex with glycerol [Thermotoga neapolitana DSM 4359],2X41_A Structure of beta-glucosidase 3B from Thermotoga neapolitana in complex with glucose [Thermotoga neapolitana DSM 4359] |
| 7MS2_A | 6.56e-46 | 1033 | 1304 | 29 | 286 | ChainA, Thermostable beta-glucosidase B [Acetivibrio thermocellus AD2],7MS2_B Chain B, Thermostable beta-glucosidase B [Acetivibrio thermocellus AD2] |
| 5WAB_A | 6.02e-41 | 1018 | 1304 | 15 | 285 | CrystalStructure of Bifidobacterium adolescentis GH3 beta-glucosidase [Bifidobacterium adolescentis ATCC 15703],5WAB_B Crystal Structure of Bifidobacterium adolescentis GH3 beta-glucosidase [Bifidobacterium adolescentis ATCC 15703],5WAB_C Crystal Structure of Bifidobacterium adolescentis GH3 beta-glucosidase [Bifidobacterium adolescentis ATCC 15703],5WAB_D Crystal Structure of Bifidobacterium adolescentis GH3 beta-glucosidase [Bifidobacterium adolescentis ATCC 15703] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P16084 | 1.08e-102 | 456 | 1271 | 30 | 793 | Beta-glucosidase A OS=Butyrivibrio fibrisolvens OX=831 GN=bglA PE=3 SV=1 |
| P15885 | 2.24e-99 | 456 | 1306 | 11 | 764 | Beta-glucosidase OS=Ruminococcus albus OX=1264 PE=3 SV=1 |
| P14002 | 3.59e-45 | 1033 | 1304 | 29 | 286 | Thermostable beta-glucosidase B OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=bglB PE=1 SV=2 |
| B8NDE2 | 9.01e-42 | 1033 | 1304 | 30 | 283 | Probable beta-glucosidase I OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=bglI PE=3 SV=2 |
| Q2U8Y5 | 9.01e-42 | 1033 | 1304 | 30 | 283 | Probable beta-glucosidase I OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=bglI PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
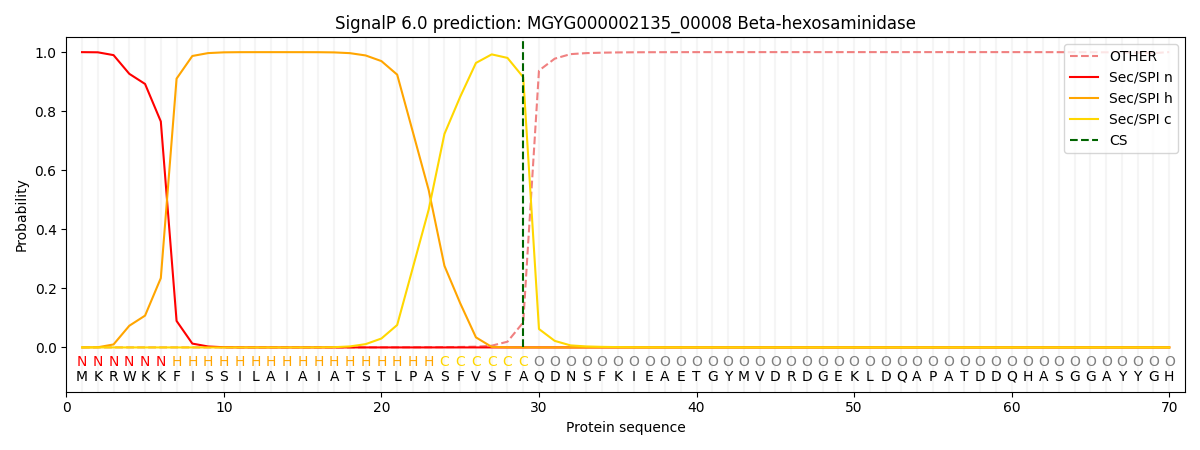
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.004772 | 0.992541 | 0.000886 | 0.000592 | 0.000604 | 0.000578 |