You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002920_00503
You are here: Home > Sequence: MGYG000002920_00503
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-485 sp900550645 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-485; CAG-485 sp900550645 | |||||||||||
| CAZyme ID | MGYG000002920_00503 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Beta-hexosaminidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 13622; End: 14773 Strand: + | |||||||||||
Full Sequence Download help
| MVWRTMTLLL LPLLAVTVAC SSPHRPSSDR TEAAAVDSAA VADSLLRVRA DSAVRALTPE | 60 |
| QRAGMLLMPA MFTRHDAPTL RQLRHYALDT HAGGIVLLRG DTLSARIIAD TLRSMGRADM | 120 |
| ILSIDAEWGL GMRLEDAPSY PVNGELATYA SERLMRSYGS RVAQQCRRLG INMVLGPVVD | 180 |
| VVPEGGQSIM SRRSFGADPA RVARLASAYA RALEEGGVIS VAKHFPGHGS VPEDSHRTLP | 240 |
| VLYKSLHLLQ SQDLLPFRIY VDSGMSGIMV GHIAVPAVDP DIQPAAVSAA VITDLLRNDM | 300 |
| NFQGLILTDA LNMGGAEGAP AWKAVAAGAD LVLAPADTDA ARRDILAAIE RGDLTQAEVD | 360 |
| AHCRRIFYYI YKLRSPRRRC AAE | 383 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 119 | 333 | 9.1e-51 | 0.9490740740740741 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1472 | BglX | 2.96e-58 | 57 | 373 | 1 | 315 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| pfam00933 | Glyco_hydro_3 | 3.47e-55 | 58 | 366 | 1 | 313 | Glycosyl hydrolase family 3 N terminal domain. |
| PRK05337 | PRK05337 | 5.44e-36 | 155 | 333 | 81 | 278 | beta-hexosaminidase; Provisional |
| PRK15098 | PRK15098 | 7.28e-09 | 170 | 366 | 150 | 349 | beta-glucosidase BglX. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCD35538.1 | 5.03e-71 | 51 | 373 | 43 | 367 |
| QUB43560.1 | 3.85e-70 | 51 | 373 | 45 | 368 |
| AOH41095.1 | 3.37e-68 | 51 | 371 | 45 | 371 |
| AVV52804.1 | 3.37e-68 | 51 | 371 | 45 | 371 |
| QUT89657.1 | 3.19e-67 | 42 | 377 | 33 | 373 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6K5J_A | 3.58e-41 | 55 | 366 | 9 | 333 | Structureof a glycoside hydrolase family 3 beta-N-acetylglucosaminidase from Paenibacillus sp. str. FPU-7 [Paenibacillaceae] |
| 3SQL_A | 6.84e-41 | 88 | 367 | 54 | 343 | CrystalStructure of Glycoside Hydrolase from Synechococcus [Synechococcus sp. PCC 7002],3SQL_B Crystal Structure of Glycoside Hydrolase from Synechococcus [Synechococcus sp. PCC 7002],3SQM_A Crystal Structure of Glycoside Hydrolase from Synechococcus Complexed with N-acetyl-D-glucosamine [Synechococcus sp. PCC 7002],3SQM_B Crystal Structure of Glycoside Hydrolase from Synechococcus Complexed with N-acetyl-D-glucosamine [Synechococcus sp. PCC 7002],3SQM_C Crystal Structure of Glycoside Hydrolase from Synechococcus Complexed with N-acetyl-D-glucosamine [Synechococcus sp. PCC 7002],3SQM_D Crystal Structure of Glycoside Hydrolase from Synechococcus Complexed with N-acetyl-D-glucosamine [Synechococcus sp. PCC 7002] |
| 6JTI_A | 2.34e-31 | 88 | 333 | 65 | 323 | Crystalstructure of native NagZ from Neisseria gonorrhoeae [Neisseria gonorrhoeae],6JTI_B Crystal structure of native NagZ from Neisseria gonorrhoeae [Neisseria gonorrhoeae],6JTI_C Crystal structure of native NagZ from Neisseria gonorrhoeae [Neisseria gonorrhoeae],6JTI_D Crystal structure of native NagZ from Neisseria gonorrhoeae [Neisseria gonorrhoeae],6JTI_E Crystal structure of native NagZ from Neisseria gonorrhoeae [Neisseria gonorrhoeae],6JTI_F Crystal structure of native NagZ from Neisseria gonorrhoeae [Neisseria gonorrhoeae],6JTJ_A Crystal structure of NagZ from Neisseria gonorrhoeae in complex with N-acetylglucosamine [Neisseria gonorrhoeae],6JTJ_B Crystal structure of NagZ from Neisseria gonorrhoeae in complex with N-acetylglucosamine [Neisseria gonorrhoeae],6JTJ_C Crystal structure of NagZ from Neisseria gonorrhoeae in complex with N-acetylglucosamine [Neisseria gonorrhoeae],6JTJ_D Crystal structure of NagZ from Neisseria gonorrhoeae in complex with N-acetylglucosamine [Neisseria gonorrhoeae],6JTJ_E Crystal structure of NagZ from Neisseria gonorrhoeae in complex with N-acetylglucosamine [Neisseria gonorrhoeae],6JTJ_F Crystal structure of NagZ from Neisseria gonorrhoeae in complex with N-acetylglucosamine [Neisseria gonorrhoeae],6JTK_A Crystal structure of NagZ from Neisseria gonorrhoeae in complex with N-trifluoroacetyl-D-glucosamine [Neisseria gonorrhoeae],6JTK_B Crystal structure of NagZ from Neisseria gonorrhoeae in complex with N-trifluoroacetyl-D-glucosamine [Neisseria gonorrhoeae],6JTK_C Crystal structure of NagZ from Neisseria gonorrhoeae in complex with N-trifluoroacetyl-D-glucosamine [Neisseria gonorrhoeae],6JTK_D Crystal structure of NagZ from Neisseria gonorrhoeae in complex with N-trifluoroacetyl-D-glucosamine [Neisseria gonorrhoeae],6JTK_E Crystal structure of NagZ from Neisseria gonorrhoeae in complex with N-trifluoroacetyl-D-glucosamine [Neisseria gonorrhoeae],6JTK_F Crystal structure of NagZ from Neisseria gonorrhoeae in complex with N-trifluoroacetyl-D-glucosamine [Neisseria gonorrhoeae],6JTL_A Crystal structure of NagZ from Neisseria gonorrhoeae in complex with zinc ion [Neisseria gonorrhoeae],6JTL_B Crystal structure of NagZ from Neisseria gonorrhoeae in complex with zinc ion [Neisseria gonorrhoeae] |
| 3BMX_A | 3.23e-31 | 48 | 366 | 33 | 389 | Beta-N-hexosaminidase(YbbD) from Bacillus subtilis [Bacillus subtilis],3BMX_B Beta-N-hexosaminidase (YbbD) from Bacillus subtilis [Bacillus subtilis],3NVD_A Structure of YBBD in complex with pugnac [Bacillus subtilis],3NVD_B Structure of YBBD in complex with pugnac [Bacillus subtilis] |
| 5BZA_A | 8.14e-31 | 94 | 313 | 31 | 249 | Crystalstructure of CbsA from Thermotoga neapolitana [Thermotoga neapolitana],5BZA_B Crystal structure of CbsA from Thermotoga neapolitana [Thermotoga neapolitana],5BZA_C Crystal structure of CbsA from Thermotoga neapolitana [Thermotoga neapolitana],5BZA_D Crystal structure of CbsA from Thermotoga neapolitana [Thermotoga neapolitana],5C0Q_A Crystal structure of Zn bound CbsA from Thermotoga neapolitana [Thermotoga neapolitana],5C0Q_B Crystal structure of Zn bound CbsA from Thermotoga neapolitana [Thermotoga neapolitana],5C0Q_C Crystal structure of Zn bound CbsA from Thermotoga neapolitana [Thermotoga neapolitana],5C0Q_D Crystal structure of Zn bound CbsA from Thermotoga neapolitana [Thermotoga neapolitana] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9JVT3 | 2.73e-32 | 88 | 333 | 29 | 287 | Beta-hexosaminidase OS=Neisseria meningitidis serogroup A / serotype 4A (strain DSM 15465 / Z2491) OX=122587 GN=nagZ PE=3 SV=1 |
| Q9K0Q4 | 1.93e-31 | 88 | 333 | 29 | 287 | Beta-hexosaminidase OS=Neisseria meningitidis serogroup B (strain MC58) OX=122586 GN=nagZ PE=3 SV=1 |
| Q7NWB7 | 2.48e-31 | 93 | 340 | 31 | 293 | Beta-hexosaminidase OS=Chromobacterium violaceum (strain ATCC 12472 / DSM 30191 / JCM 1249 / NBRC 12614 / NCIMB 9131 / NCTC 9757) OX=243365 GN=nagZ PE=3 SV=1 |
| A9M1Z4 | 2.67e-31 | 88 | 333 | 29 | 287 | Beta-hexosaminidase OS=Neisseria meningitidis serogroup C (strain 053442) OX=374833 GN=nagZ PE=3 SV=1 |
| Q5FA94 | 7.08e-31 | 88 | 333 | 29 | 287 | Beta-hexosaminidase OS=Neisseria gonorrhoeae (strain ATCC 700825 / FA 1090) OX=242231 GN=nagZ PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
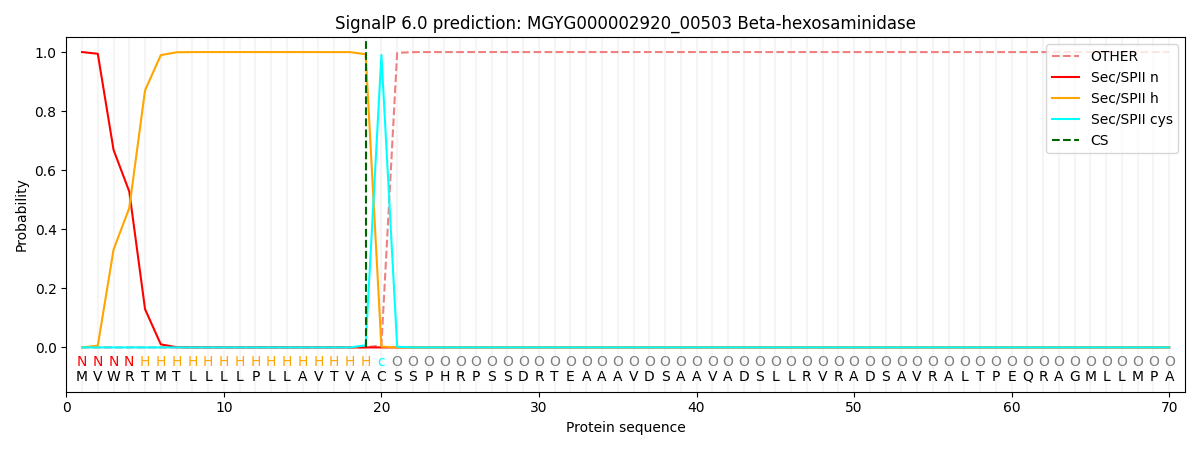
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000101 | 0.000000 | 0.000000 | 0.000000 |
