You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003172_00039
You are here: Home > Sequence: MGYG000003172_00039
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-485 sp900555915 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-485; CAG-485 sp900555915 | |||||||||||
| CAZyme ID | MGYG000003172_00039 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Xylan 1,4-beta-xylosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3666; End: 6593 Strand: + | |||||||||||
Full Sequence Download help
| MKKYCLPVIL SAVLMLTSAG KQKEVSFDSY NHDFGTVNSK QGAVSHRFRL SNNGKTPVTV | 60 |
| ARTSPSCDCI QALIPESAIE PGQSADILVV LDPSGNVGKS IRSVEIIDIN GKTLGALSVE | 120 |
| SDIEEAPFPG NYPFRNTGLG VEERVENLIS LLTPEEKVGL MMNKSVSVDR LGIPSYNWWS | 180 |
| EACHGIRQDG YTVFPQPIGM ASAFNPALVY DVFSAVSDEA RANWNRSDHD IYGVGMGEIY | 240 |
| YPGNPELTFW CPNVNIFRDP RWGRGQETCG EDPYLNAVLG VETVRGMQGD DSRYFKTHAC | 300 |
| AKHYAVHSGP EPLRHTYNAS VSMRDLWETY LPAFKALVKK GDVREVMCAY NRYEGKPCCM | 360 |
| SDRLLIDILR RKWGFDGIIL TDCDAINNFY NKGQHETHSD PMSASVDAVL NGTDLECGKV | 420 |
| FMSLTEALEK GKIEEKDLDA HLRRTLKGRF ELGMFDPAHM LPWSGMQEAV ISSEPFHQLA | 480 |
| VQAARESIVL LDNDGVLPLD SNVKKIAVVG PNADDTSMLN GNYGGTPTAA HTSSLLDGIR | 540 |
| DAFPSAEIIY RKGCELDDDY RTIPYMKDFN DGKGLLVEFF NNSDLSGDPA VTAYHDRLNF | 600 |
| STFGAWGFAE GISKDTLSVR VSGLWKAPFT GDMSYNITTD NGYILKINGK TVEDTSGQAG | 660 |
| RRGFGFNRKI DYRQYPVIEG DTYDICIEYR HGASNFAMLR GELCRRQLVE FDDIASEVKD | 720 |
| ADAIIVIGGI SAQMEGEGGD KDDIELPAVQ QRLLKAMHST GRPVIMVNCS GSAIAFGSVE | 780 |
| GDYDALIQAW YPGQGGSQAL AEVISGAYNP GGKLPVTFYR SNDDLPDFLD YSMDNRTYRY | 840 |
| FKGKPQYAFG YGLSYTDFEI GKGKLSRKSM KKDGKVTLTV PVTNTGKRHG TETVQAYIEA | 900 |
| LDYDDAPLKS LKGFSKISLA PGESGRAVIT LDAEAFEFYD PSVDELSPRS GRYRILYGSS | 960 |
| SRDEDLRALE FTLKD | 975 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 167 | 417 | 3.1e-66 | 0.9814814814814815 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PLN03080 | PLN03080 | 9.24e-116 | 127 | 932 | 35 | 743 | Probable beta-xylosidase; Provisional |
| PRK15098 | PRK15098 | 4.82e-103 | 161 | 973 | 92 | 764 | beta-glucosidase BglX. |
| COG1472 | BglX | 1.78e-68 | 161 | 543 | 48 | 387 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| pfam01915 | Glyco_hydro_3_C | 2.27e-44 | 488 | 855 | 1 | 216 | Glycosyl hydrolase family 3 C-terminal domain. This domain is involved in catalysis and may be involved in binding beta-glucan. This domain is found associated with pfam00933. |
| pfam00933 | Glyco_hydro_3 | 1.86e-38 | 161 | 446 | 54 | 314 | Glycosyl hydrolase family 3 N terminal domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCD36993.1 | 0.0 | 88 | 974 | 1 | 889 |
| QVJ80234.1 | 0.0 | 132 | 974 | 23 | 869 |
| ADE83440.1 | 0.0 | 132 | 974 | 23 | 869 |
| QRQ48937.1 | 0.0 | 132 | 970 | 28 | 880 |
| QUT44634.1 | 0.0 | 132 | 970 | 28 | 880 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7VC7_A | 4.13e-84 | 144 | 955 | 25 | 715 | ChainA, xylan 1,4-beta-xylosidase [Phanerodontia chrysosporium] |
| 7VC6_A | 4.13e-84 | 144 | 955 | 25 | 715 | ChainA, xylan 1,4-beta-xylosidase [Phanerodontia chrysosporium] |
| 5A7M_A | 2.08e-76 | 108 | 973 | 16 | 753 | ChainA, BETA-XYLOSIDASE [Trichoderma reesei],5A7M_B Chain B, BETA-XYLOSIDASE [Trichoderma reesei] |
| 5AE6_A | 2.12e-76 | 108 | 973 | 16 | 753 | ChainA, BETA-XYLOSIDASE [Trichoderma reesei],5AE6_B Chain B, BETA-XYLOSIDASE [Trichoderma reesei] |
| 6Q7I_A | 4.18e-73 | 133 | 935 | 41 | 718 | GH3exo-beta-xylosidase (XlnD) [Aspergillus nidulans FGSC A4],6Q7I_B GH3 exo-beta-xylosidase (XlnD) [Aspergillus nidulans FGSC A4],6Q7J_A Chain A, Exo-1,4-beta-xylosidase xlnD [Aspergillus nidulans FGSC A4],6Q7J_B Chain B, Exo-1,4-beta-xylosidase xlnD [Aspergillus nidulans FGSC A4] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| D5EY15 | 5.28e-230 | 133 | 973 | 23 | 861 | Xylan 1,4-beta-xylosidase OS=Prevotella ruminicola (strain ATCC 19189 / JCM 8958 / 23) OX=264731 GN=xyl3A PE=1 SV=1 |
| Q94KD8 | 3.83e-94 | 134 | 931 | 44 | 723 | Probable beta-D-xylosidase 2 OS=Arabidopsis thaliana OX=3702 GN=BXL2 PE=2 SV=1 |
| A5JTQ2 | 2.24e-93 | 122 | 932 | 42 | 738 | Beta-xylosidase/alpha-L-arabinofuranosidase 1 (Fragment) OS=Medicago sativa subsp. varia OX=36902 GN=Xyl1 PE=1 SV=1 |
| Q9FLG1 | 2.79e-93 | 122 | 949 | 47 | 762 | Beta-D-xylosidase 4 OS=Arabidopsis thaliana OX=3702 GN=BXL4 PE=1 SV=1 |
| A5JTQ3 | 5.96e-93 | 122 | 919 | 42 | 725 | Beta-xylosidase/alpha-L-arabinofuranosidase 2 OS=Medicago sativa subsp. varia OX=36902 GN=Xyl2 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
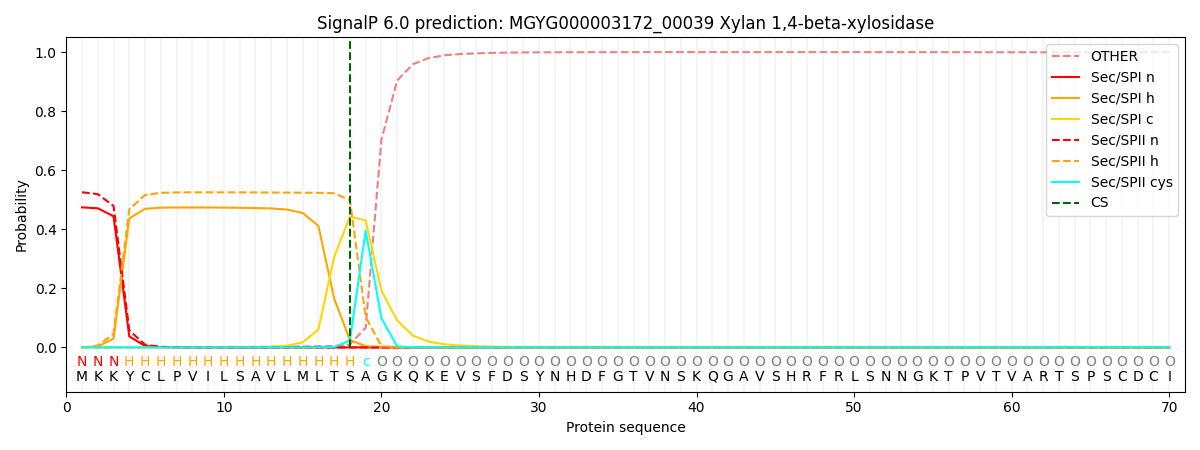
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001338 | 0.468555 | 0.529460 | 0.000279 | 0.000188 | 0.000160 |
