You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003411_00026
You are here: Home > Sequence: MGYG000003411_00026
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
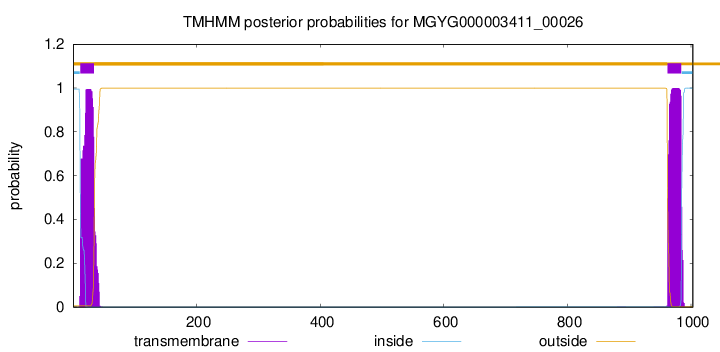
TMHMM annotations
Basic Information help
| Species | CAG-568 sp000434395 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; RFN20; CAG-288; CAG-568; CAG-568 sp000434395 | |||||||||||
| CAZyme ID | MGYG000003411_00026 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 33558; End: 36569 Strand: + | |||||||||||
Full Sequence Download help
| MMIKKLWKKN SFKIWSITSG VLLATVIAVP IVLNTALADV LDFALGGADP IFKEGEQSAY | 60 |
| VKSFNSKDES KANADNVNVK LNEEGMVLLK NKNNALPLAK GSKISVFGKN SDNIAIGGSG | 120 |
| SGAASGDFIG VNESLRNAGF EVNPTLENFY HDNKLSGEGR SSASGDLDSG KPVTLKTGET | 180 |
| PQSSYTNEVK DSYKNYQDAA IIVLTRIGGE GMDLPMTMDG VEGARKSDDH YLQLDKNEED | 240 |
| LIVAVTNGGF KKVIVVINSG SAMELGFLEE NAKYVTQKGY TIDPSKIDAA ISIGYPGNSG | 300 |
| CKALGEIIAG DVNPSGKTVD TYATDFKKMP SWKNFSVNND FDTEGGLGDQ YMIGDQAKLY | 360 |
| YFADYEEGEY VGYRYYETRG FTDGNEWYND NVVYPFGYGL SYTTFSWSVD ATTVPSTLDP | 420 |
| TNEFTVEVEV KNEGSVAGKD VVEVYAKAPY KTGEIEKPYE ILIGYAKTGL LEPGASEKVS | 480 |
| IDINPYYMAS YDYKDANNNG FKGFELDAGT YSLSFNKNSH ESIRSFDMEL ADNYQYAEDP | 540 |
| VTGTEVKNLY TDNVDASFNS DYQLKTILSR KDWEGTFPSP RTQEERTVSQ DFIDKLDDKS | 600 |
| TNNPNDYDND PQFDYPTVDD TKLYTIYDLL YKDNLDADGN YVVDEEGNKV RGEWVGEVSF | 660 |
| DDERWDRLLD QMNLAEVANM YNTAAYKIAG VDSVGLPVVT CADGPVGWTS FINPGLFEKT | 720 |
| ASYSCGVTVA STWNDELISE FGAAVGEEGL IGNGSTPYTG WYAPAMNIHR SPFGGRNFEY | 780 |
| FSEDPLLSGK VGAAEVRGCR SKGLVTFVKH FAANEQETHR SISGDCSWIT EQALREIYLR | 840 |
| PFEITVKEGK TLGMMSSFNR IGTRWTGGDY RLLTTILRDE WGFNGAVICD FNTIPTYMDA | 900 |
| RQEAYAGGDL NLATMASSAW NYDESSIGDQ LVLRRVLKDV CYSIVNANSM QGTVIGQTAP | 960 |
| KWFWINYAYI FGMLGVILVW GGIVITLDIK KIKAEEKAES DSK | 1003 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 690 | 912 | 1.9e-48 | 0.9814814814814815 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1472 | BglX | 1.24e-28 | 677 | 890 | 41 | 248 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| pfam01915 | Glyco_hydro_3_C | 8.42e-23 | 86 | 373 | 1 | 216 | Glycosyl hydrolase family 3 C-terminal domain. This domain is involved in catalysis and may be involved in binding beta-glucan. This domain is found associated with pfam00933. |
| PRK15098 | PRK15098 | 6.64e-22 | 84 | 493 | 393 | 739 | beta-glucosidase BglX. |
| pfam00933 | Glyco_hydro_3 | 6.68e-20 | 730 | 890 | 95 | 254 | Glycosyl hydrolase family 3 N terminal domain. |
| PLN03080 | PLN03080 | 2.81e-17 | 84 | 521 | 401 | 774 | Probable beta-xylosidase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| VEU80232.1 | 1.16e-274 | 5 | 993 | 7 | 961 |
| QOS39239.1 | 8.87e-267 | 6 | 991 | 3 | 965 |
| VEU80230.1 | 1.08e-204 | 80 | 914 | 45 | 860 |
| QQA37675.1 | 3.02e-175 | 73 | 962 | 88 | 892 |
| QOL32307.1 | 1.02e-174 | 2 | 965 | 14 | 938 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5WUG_A | 2.29e-71 | 84 | 891 | 46 | 748 | Expression,characterization and crystal structure of a novel beta-glucosidase from Paenibacillus barengoltzii [Paenibacillus barengoltzii],5WVP_A Expression, characterization and crystal structure of a novel beta-glucosidase from Paenibacillus barengoltzii [Paenibacillus barengoltzii] |
| 2X40_A | 1.74e-41 | 663 | 891 | 2 | 243 | Structureof beta-glucosidase 3B from Thermotoga neapolitana in complex with glycerol [Thermotoga neapolitana DSM 4359],2X41_A Structure of beta-glucosidase 3B from Thermotoga neapolitana in complex with glucose [Thermotoga neapolitana DSM 4359] |
| 2X42_A | 9.79e-41 | 663 | 888 | 2 | 240 | Structureof beta-glucosidase 3B from Thermotoga neapolitana in complex with alpha-D-glucose [Thermotoga neapolitana DSM 4359] |
| 5WAB_A | 3.80e-32 | 669 | 912 | 9 | 249 | CrystalStructure of Bifidobacterium adolescentis GH3 beta-glucosidase [Bifidobacterium adolescentis ATCC 15703],5WAB_B Crystal Structure of Bifidobacterium adolescentis GH3 beta-glucosidase [Bifidobacterium adolescentis ATCC 15703],5WAB_C Crystal Structure of Bifidobacterium adolescentis GH3 beta-glucosidase [Bifidobacterium adolescentis ATCC 15703],5WAB_D Crystal Structure of Bifidobacterium adolescentis GH3 beta-glucosidase [Bifidobacterium adolescentis ATCC 15703] |
| 3AC0_A | 1.01e-29 | 666 | 896 | 7 | 232 | Crystalstructure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus],3AC0_B Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus],3AC0_C Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus],3AC0_D Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P16084 | 3.81e-83 | 84 | 912 | 38 | 788 | Beta-glucosidase A OS=Butyrivibrio fibrisolvens OX=831 GN=bglA PE=3 SV=1 |
| P15885 | 2.97e-65 | 81 | 946 | 16 | 782 | Beta-glucosidase OS=Ruminococcus albus OX=1264 PE=3 SV=1 |
| P27034 | 1.47e-35 | 666 | 891 | 3 | 223 | Beta-glucosidase OS=Rhizobium radiobacter OX=358 GN=cbg-1 PE=3 SV=1 |
| Q5BFG8 | 2.50e-33 | 668 | 912 | 20 | 249 | Beta-glucosidase B OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=bglB PE=1 SV=1 |
| B8NDE2 | 5.66e-33 | 666 | 961 | 7 | 294 | Probable beta-glucosidase I OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=bglI PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
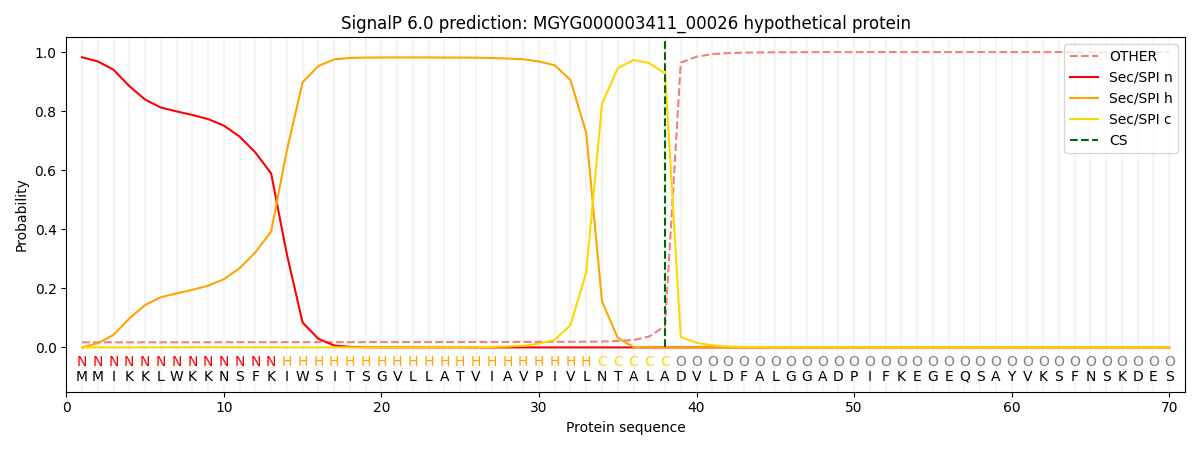
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.020284 | 0.978594 | 0.000387 | 0.000251 | 0.000225 | 0.000243 |