You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003630_00327
You are here: Home > Sequence: MGYG000003630_00327
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | F082 sp900771045 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; F082; F082; F082 sp900771045 | |||||||||||
| CAZyme ID | MGYG000003630_00327 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Beta-hexosaminidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1373; End: 4252 Strand: + | |||||||||||
Full Sequence Download help
| MRRLFFVLSF FIIANQIFAQ KQQEREWVDS VYMSLTLEQK VGQLMNLRAN KSSQPYFPLI | 60 |
| DEMIRKYDIG GVTFFRYGDA KSQLEQTIKW QSEAQTPLMI TIDGEWGLGM RLRDCISYPY | 120 |
| QMTLGALRED DLIAEMGSQI AEQCRMMGIQ SSFSPVVDVN SEPSNPVIGM RSFGENPEKV | 180 |
| ARKGLVYARA LQNNGILPTM KHFPGHGDTH TDSHVALPIV AKSLSKIEKN ELVPFKHLID | 240 |
| NGISGLMVGH LSYPAIDDSG TPTSLSVKTL DGFLRKELGF EGLIVTDGLD MKGLTGGNDP | 300 |
| DKACLMALMA GADVLLLPVN PEASIKIIVE AAKSDDDVMR RVSESCRRVL TWKYRLGLAE | 360 |
| KQSLEPSEIS SKMKNPRYAE LKQRIYDEAV TVLSNSDFTP LNKDEKMAFI STISDDNMYF | 420 |
| LLKNKGFKCE KILFENCCQN LKETAKKLKD YDRVIVNVRN TSVFAKKHYG INQETIDLVN | 480 |
| ALSKNKNLIL NLFTLPYAID MFDFAKKTTD IVVGYEDNVM VERSVTAVLC GDIASRGVLP | 540 |
| VSTRLYKEGS GAAYNNDVKS IDNEDVRMDL VFERKVDSIV LEGLGQQAYP GCQVLAVKDG | 600 |
| VVIYDKCFGT FTYDDDRAVQ PDDVYDIASL TKIFGSTIAI MKLYDENKID INAKLSDFFP | 660 |
| FLRGTNKADM TLLEIMTHQS GMQAWHPFYL ETIENGMLNP LIYSDYIDED HPLCVAEDVY | 720 |
| IGKNYWRDML EAFVATEMGE KKYKYCDMGF YFMPKIVELT TGVPFEEYLD ETFYRPMGLS | 780 |
| HIFFNPLKHI EIDKIAPTAY DSVFRKQFLR GRVHDQAAAM MGGISGHAGL FANAYDLAAI | 840 |
| MQMLVDGGVY NGRRYLSKAT VKMFTTAPFT DNDNRRGIGF DKPNIDPDCP FTTPSLRASM | 900 |
| KSFGHTGFTG TFAWADPENK LVVIFLSNRV CPDDNNNKLS DMNIRTNIHD VFYQMEEEK | 959 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 91 | 316 | 9.1e-55 | 0.9768518518518519 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00933 | Glyco_hydro_3 | 5.10e-79 | 36 | 351 | 1 | 315 | Glycosyl hydrolase family 3 N terminal domain. |
| COG1472 | BglX | 4.71e-75 | 35 | 417 | 1 | 367 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| PRK03642 | PRK03642 | 2.56e-48 | 567 | 937 | 42 | 408 | putative periplasmic esterase; Provisional |
| pfam00144 | Beta-lactamase | 1.88e-47 | 576 | 929 | 1 | 308 | Beta-lactamase. This family appears to be distantly related to pfam00905 and PF00768 D-alanyl-D-alanine carboxypeptidase. |
| COG1680 | AmpC | 2.77e-45 | 590 | 938 | 54 | 372 | CubicO group peptidase, beta-lactamase class C family [Defense mechanisms]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QHT66932.1 | 4.88e-233 | 22 | 954 | 53 | 997 |
| CAG5080436.1 | 8.02e-228 | 22 | 953 | 47 | 993 |
| ATL46696.1 | 1.15e-224 | 27 | 954 | 60 | 1001 |
| QIA09613.1 | 3.53e-222 | 23 | 953 | 33 | 976 |
| APS38154.1 | 7.85e-221 | 26 | 953 | 39 | 978 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3BMX_A | 1.41e-65 | 8 | 541 | 15 | 615 | Beta-N-hexosaminidase(YbbD) from Bacillus subtilis [Bacillus subtilis],3BMX_B Beta-N-hexosaminidase (YbbD) from Bacillus subtilis [Bacillus subtilis],3NVD_A Structure of YBBD in complex with pugnac [Bacillus subtilis],3NVD_B Structure of YBBD in complex with pugnac [Bacillus subtilis] |
| 3LK6_A | 3.66e-64 | 35 | 541 | 16 | 589 | ChainA, Lipoprotein ybbD [Bacillus subtilis],3LK6_B Chain B, Lipoprotein ybbD [Bacillus subtilis],3LK6_C Chain C, Lipoprotein ybbD [Bacillus subtilis],3LK6_D Chain D, Lipoprotein ybbD [Bacillus subtilis] |
| 4GYJ_A | 4.98e-64 | 11 | 541 | 22 | 619 | Crystalstructure of mutant (D318N) bacillus subtilis family 3 glycoside hydrolase (nagz) in complex with glcnac-murnac (space group P1) [Bacillus subtilis subsp. subtilis str. 168],4GYJ_B Crystal structure of mutant (D318N) bacillus subtilis family 3 glycoside hydrolase (nagz) in complex with glcnac-murnac (space group P1) [Bacillus subtilis subsp. subtilis str. 168],4GYK_A Crystal structure of mutant (D318N) bacillus subtilis family 3 glycoside hydrolase (nagz) in complex with glcnac-murnac (space group P1211) [Bacillus subtilis subsp. subtilis str. 168],4GYK_B Crystal structure of mutant (D318N) bacillus subtilis family 3 glycoside hydrolase (nagz) in complex with glcnac-murnac (space group P1211) [Bacillus subtilis subsp. subtilis str. 168] |
| 6K5J_A | 3.11e-58 | 34 | 540 | 10 | 528 | Structureof a glycoside hydrolase family 3 beta-N-acetylglucosaminidase from Paenibacillus sp. str. FPU-7 [Paenibacillaceae] |
| 3SQL_A | 9.84e-53 | 69 | 370 | 58 | 362 | CrystalStructure of Glycoside Hydrolase from Synechococcus [Synechococcus sp. PCC 7002],3SQL_B Crystal Structure of Glycoside Hydrolase from Synechococcus [Synechococcus sp. PCC 7002],3SQM_A Crystal Structure of Glycoside Hydrolase from Synechococcus Complexed with N-acetyl-D-glucosamine [Synechococcus sp. PCC 7002],3SQM_B Crystal Structure of Glycoside Hydrolase from Synechococcus Complexed with N-acetyl-D-glucosamine [Synechococcus sp. PCC 7002],3SQM_C Crystal Structure of Glycoside Hydrolase from Synechococcus Complexed with N-acetyl-D-glucosamine [Synechococcus sp. PCC 7002],3SQM_D Crystal Structure of Glycoside Hydrolase from Synechococcus Complexed with N-acetyl-D-glucosamine [Synechococcus sp. PCC 7002] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P40406 | 7.75e-65 | 8 | 541 | 15 | 615 | Beta-hexosaminidase OS=Bacillus subtilis (strain 168) OX=224308 GN=nagZ PE=1 SV=1 |
| P48823 | 5.94e-53 | 58 | 478 | 53 | 496 | Beta-hexosaminidase A OS=Pseudoalteromonas piscicida OX=43662 GN=cht60 PE=1 SV=1 |
| Q8XBJ0 | 9.21e-42 | 520 | 929 | 5 | 395 | Putative D-alanyl-D-alanine carboxypeptidase OS=Escherichia coli O157:H7 OX=83334 GN=yfeW PE=3 SV=1 |
| P77619 | 5.65e-41 | 589 | 929 | 55 | 395 | Putative D-alanyl-D-alanine carboxypeptidase OS=Escherichia coli (strain K12) OX=83333 GN=yfeW PE=1 SV=2 |
| Q0AF74 | 7.06e-39 | 69 | 328 | 28 | 293 | Beta-hexosaminidase OS=Nitrosomonas eutropha (strain DSM 101675 / C91 / Nm57) OX=335283 GN=nagZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
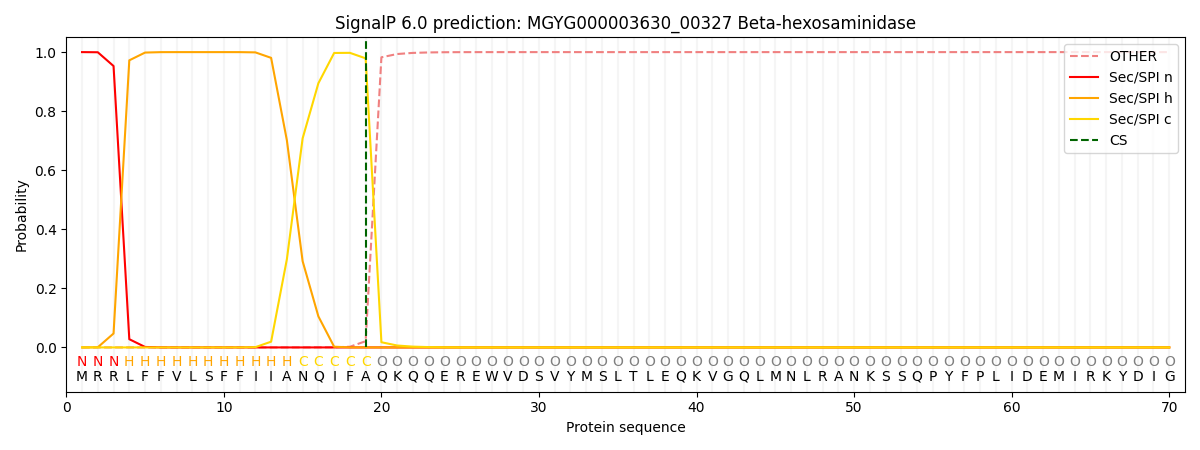
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000213 | 0.999155 | 0.000155 | 0.000152 | 0.000146 | 0.000136 |
