You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003932_00184
You are here: Home > Sequence: MGYG000003932_00184
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
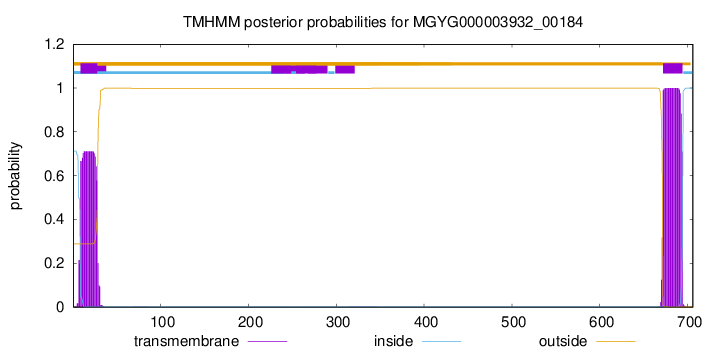
TMHMM annotations
Basic Information help
| Species | CAG-964 sp000435335 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Acutalibacteraceae; CAG-964; CAG-964 sp000435335 | |||||||||||
| CAZyme ID | MGYG000003932_00184 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Beta-hexosaminidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 207773; End: 209893 Strand: + | |||||||||||
Full Sequence Download help
| MRTGFKKAVS VALCVLCAAV GIPTASAADN VDTLLSKMTT EEKIAQMITV AVRQWDGEDF | 60 |
| TVMEDDVANI ISKTSVGGVI LFAENLVDTE QSVKLTTAFQ QAALASKNKI PLFISTDQEG | 120 |
| GEVVRLNTGT SLPGNMALGA INSANDAKTA GNILGSELAA LGINMDFAPS LDVNSNPSNP | 180 |
| IIGLRSFSSD PAIVSALGTQ VINGIQQNNV AAAAKHFPGH GDTSTDSHTS LPSVDKSLDL | 240 |
| LEKCEFLPFK AAIDNGVDVI MTAHIQFPQV EKKTVKSESD GSNVTLPATL SKTFVTDILR | 300 |
| NKLGFKGVVT TDAMNMGAIA DNFGVAQACI LAINAGVDNL LMPISLTDKS SGTELKGLIG | 360 |
| NIKSAVVNGN IKEETIDNAV KRILQLKEKR GVLNYTAPTA ENAKKIVGSA ENHEKEDDLA | 420 |
| AKAVTVLKNE GNVLPFKPTE NQNIALVAAY ENETPGMEYA MSKMISQKTI PNVNYETCYY | 480 |
| NDGSSAEEIL NTVSGADYVI VLSEMNSNED IAQGSLNTYV PTSVMNYCKE NNIKCVVVSV | 540 |
| GKPYDTANYK DAQALMAAYG CNGMNPEDTD GSQPATYTYG PNILAAVEVA FGYKNPYGTL | 600 |
| PVDIPAVLSD GSMYESNLVF KKGDGLLFAG TVRPAEAATQ PAATEDEKAT ALPSSSDPVN | 660 |
| SFMNTLQNSG YFLIVACLLA AALVVIIIII AVAVKGGKKK SNKKIR | 706 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 105 | 342 | 7.8e-66 | 0.9814814814814815 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00933 | Glyco_hydro_3 | 7.73e-106 | 39 | 386 | 1 | 316 | Glycosyl hydrolase family 3 N terminal domain. |
| COG1472 | BglX | 1.48e-102 | 38 | 456 | 1 | 369 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| PRK05337 | PRK05337 | 1.08e-58 | 76 | 342 | 26 | 279 | beta-hexosaminidase; Provisional |
| PRK15098 | PRK15098 | 1.06e-31 | 8 | 447 | 14 | 416 | beta-glucosidase BglX. |
| pfam01915 | Glyco_hydro_3_C | 7.56e-08 | 424 | 610 | 1 | 195 | Glycosyl hydrolase family 3 C-terminal domain. This domain is involved in catalysis and may be involved in binding beta-glucan. This domain is found associated with pfam00933. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AEH55774.1 | 2.55e-180 | 24 | 628 | 82 | 694 |
| AYE33655.1 | 8.23e-180 | 21 | 626 | 24 | 642 |
| QAS61817.1 | 8.23e-180 | 21 | 626 | 24 | 642 |
| AFJ26297.1 | 8.62e-180 | 24 | 628 | 82 | 694 |
| SQF71406.1 | 2.28e-177 | 27 | 673 | 129 | 778 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3BMX_A | 2.54e-153 | 30 | 626 | 34 | 634 | Beta-N-hexosaminidase(YbbD) from Bacillus subtilis [Bacillus subtilis],3BMX_B Beta-N-hexosaminidase (YbbD) from Bacillus subtilis [Bacillus subtilis],3NVD_A Structure of YBBD in complex with pugnac [Bacillus subtilis],3NVD_B Structure of YBBD in complex with pugnac [Bacillus subtilis] |
| 3LK6_A | 6.33e-153 | 30 | 626 | 8 | 608 | ChainA, Lipoprotein ybbD [Bacillus subtilis],3LK6_B Chain B, Lipoprotein ybbD [Bacillus subtilis],3LK6_C Chain C, Lipoprotein ybbD [Bacillus subtilis],3LK6_D Chain D, Lipoprotein ybbD [Bacillus subtilis] |
| 4GYJ_A | 1.71e-152 | 30 | 626 | 38 | 638 | Crystalstructure of mutant (D318N) bacillus subtilis family 3 glycoside hydrolase (nagz) in complex with glcnac-murnac (space group P1) [Bacillus subtilis subsp. subtilis str. 168],4GYJ_B Crystal structure of mutant (D318N) bacillus subtilis family 3 glycoside hydrolase (nagz) in complex with glcnac-murnac (space group P1) [Bacillus subtilis subsp. subtilis str. 168],4GYK_A Crystal structure of mutant (D318N) bacillus subtilis family 3 glycoside hydrolase (nagz) in complex with glcnac-murnac (space group P1211) [Bacillus subtilis subsp. subtilis str. 168],4GYK_B Crystal structure of mutant (D318N) bacillus subtilis family 3 glycoside hydrolase (nagz) in complex with glcnac-murnac (space group P1211) [Bacillus subtilis subsp. subtilis str. 168] |
| 6K5J_A | 2.46e-87 | 38 | 431 | 11 | 380 | Structureof a glycoside hydrolase family 3 beta-N-acetylglucosaminidase from Paenibacillus sp. str. FPU-7 [Paenibacillaceae] |
| 4ZM6_A | 3.96e-73 | 67 | 607 | 30 | 530 | Aunique GCN5-related glucosamine N-acetyltransferase region exist in the fungal multi-domain GH3 beta-N-acetylglucosaminidase [Rhizomucor miehei CAU432],4ZM6_B A unique GCN5-related glucosamine N-acetyltransferase region exist in the fungal multi-domain GH3 beta-N-acetylglucosaminidase [Rhizomucor miehei CAU432] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P40406 | 1.39e-152 | 30 | 626 | 34 | 634 | Beta-hexosaminidase OS=Bacillus subtilis (strain 168) OX=224308 GN=nagZ PE=1 SV=1 |
| P48823 | 1.71e-87 | 38 | 604 | 16 | 596 | Beta-hexosaminidase A OS=Pseudoalteromonas piscicida OX=43662 GN=cht60 PE=1 SV=1 |
| Q7WUL3 | 3.24e-48 | 31 | 603 | 19 | 554 | Beta-N-acetylglucosaminidase/beta-glucosidase OS=Cellulomonas fimi OX=1708 GN=nag3 PE=1 SV=1 |
| Q0AF74 | 4.79e-38 | 71 | 388 | 23 | 325 | Beta-hexosaminidase OS=Nitrosomonas eutropha (strain DSM 101675 / C91 / Nm57) OX=335283 GN=nagZ PE=3 SV=1 |
| Q3SKU2 | 6.11e-38 | 65 | 342 | 17 | 282 | Beta-hexosaminidase OS=Thiobacillus denitrificans (strain ATCC 25259) OX=292415 GN=nagZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
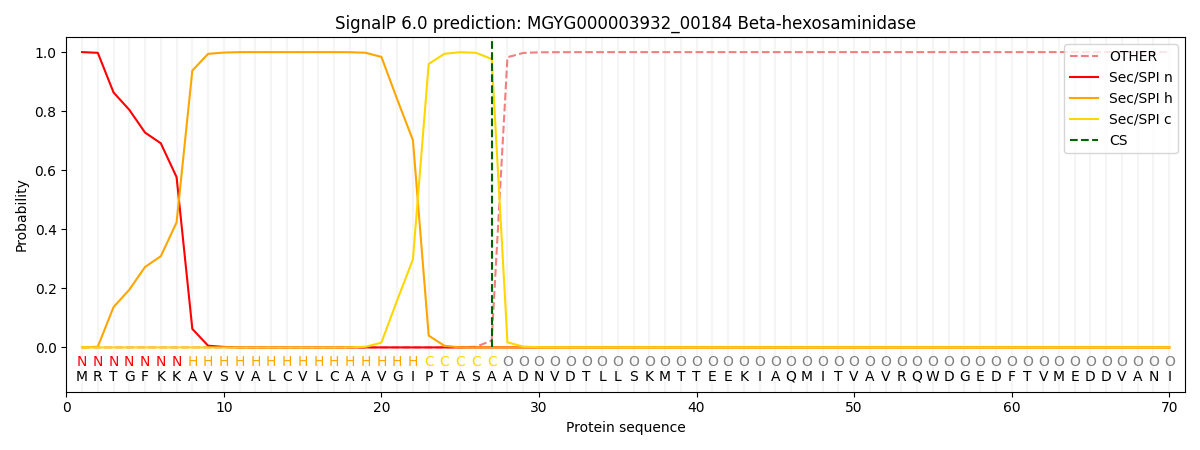
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000234 | 0.999093 | 0.000204 | 0.000161 | 0.000155 | 0.000142 |