You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000969_00460
You are here: Home > Sequence: MGYG000000969_00460
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
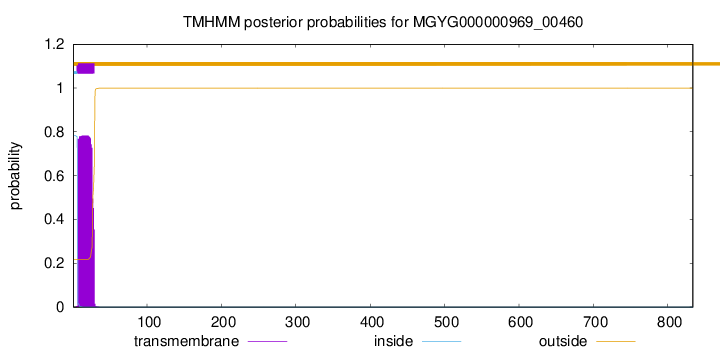
TMHMM annotations
Basic Information help
| Species | Eubacterium_I sp900546495 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Eubacterium_I; Eubacterium_I sp900546495 | |||||||||||
| CAZyme ID | MGYG000000969_00460 | |||||||||||
| CAZy Family | GH30 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 63234; End: 65738 Strand: + | |||||||||||
Full Sequence Download help
| MKRMRQFASL IAAAAMVVTS VPMNGWTVQA QAATTPTVDT TVKLKPAEAS TFNDTNKDGL | 60 |
| GEFQGWGTSL CWWANRIGYS DKLTEQAANL FFSADGLNMN IGRYNIGGGD DVADTVTTTK | 120 |
| VPVNENATFY DLAAGTYTYN GTSGKAETYT KMKDMTYSRS DAEFGITKGE KVGSFNKLGW | 180 |
| INKLDDAAGS GDNLRYNVNV EEAGEYTVKL LLTLEGTNSR DVAIRVNDDS TQDKVISSSD | 240 |
| INSSIIAEGT NNGSHCMLFC VTISGVQLKK GENTINIAGK SDWTLDFVKM AVVKTSDLAE | 300 |
| LPEEDEFLHQ SHIVRSDGGV PGYATEVTKI DTTKHDISYY ADKNNFDQYD EDCGYAWKYD | 360 |
| WTADQNQVNV LKAVAKASGN DFIAEAFSNS PPYFMTVSGC SSGNTNSSQD NLRADSVNAF | 420 |
| AKYMADVMEH WQEEGVINFQ SVDPMNEPYT DYWGANSNKQ EGCHFDQGES QSRVLVALNK | 480 |
| ELQNRESLKG VIISGTDETS IDTQITSYNK LTDEAKNVIS RIDTHTYSGS NRSGLKETAQ | 540 |
| NAGKNLWMSE VDGAYTAGSN AGEMTAALGL AQRMMTDVNG LEASAWILWN AIDMHADSSE | 600 |
| YGQKWINKGS ENDYLTMTDL EKAWKSQSSN GYWGIAAADH NNNEIVLSQK YYAYGQFSRY | 660 |
| IRPGDTIIGS DQEGKTLAAY DVDGNKAIIV AINTSSSDQT WEFDMSGFEE MGNKVTAIRT | 720 |
| SGDLKTGEHW KDVTKSDNIV VDADEQCFTA TMKGNSITTY IVEGVNGIKD TSDDNTAEKP | 780 |
| EVRQIAIAKN QVTGSAPWNN GTTNVASDVV DNNYGTFFDG VSNGYVTLDL GQET | 834 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH30 | 311 | 760 | 3e-123 | 0.8574561403508771 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam14587 | Glyco_hydr_30_2 | 1.32e-19 | 358 | 593 | 97 | 357 | O-Glycosyl hydrolase family 30. |
| COG5520 | XynC | 4.37e-08 | 390 | 676 | 124 | 374 | O-Glycosyl hydrolase [Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QBE99233.1 | 8.66e-313 | 1 | 779 | 1 | 748 |
| QMW77599.1 | 8.05e-311 | 2 | 779 | 23 | 772 |
| QIB58165.1 | 2.71e-310 | 1 | 779 | 1 | 748 |
| ASU27677.1 | 4.30e-308 | 1 | 779 | 1 | 748 |
| ANU74869.1 | 4.30e-308 | 1 | 779 | 1 | 748 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q76FP5 | 5.70e-37 | 359 | 765 | 126 | 476 | Endo-beta-1,6-galactanase OS=Hypocrea rufa OX=5547 GN=6GAL PE=1 SV=1 |
| A0A401ETL2 | 3.59e-29 | 364 | 808 | 172 | 684 | Exo-beta-1,6-galactobiohydrolase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=bl1,6Gal PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
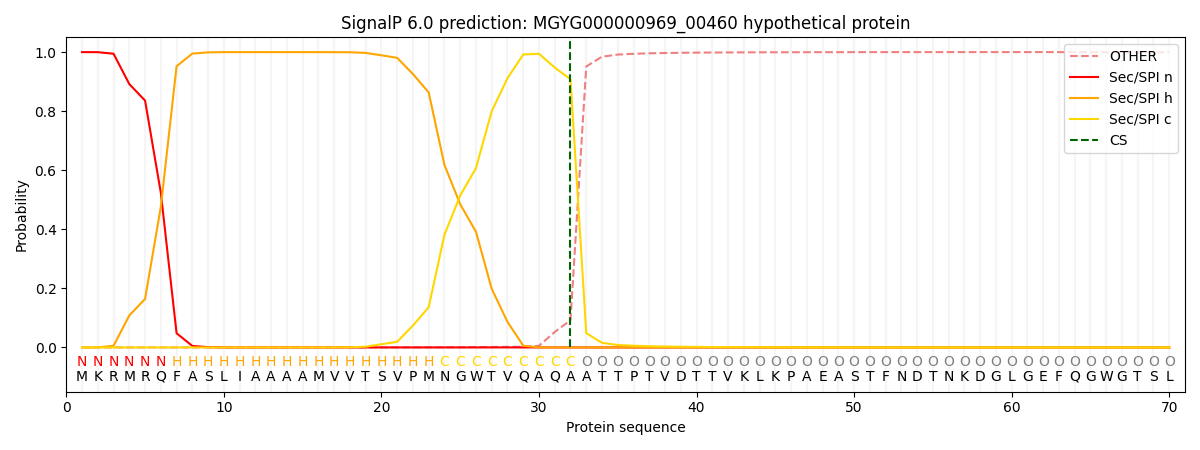
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001846 | 0.996528 | 0.000338 | 0.000508 | 0.000395 | 0.000344 |